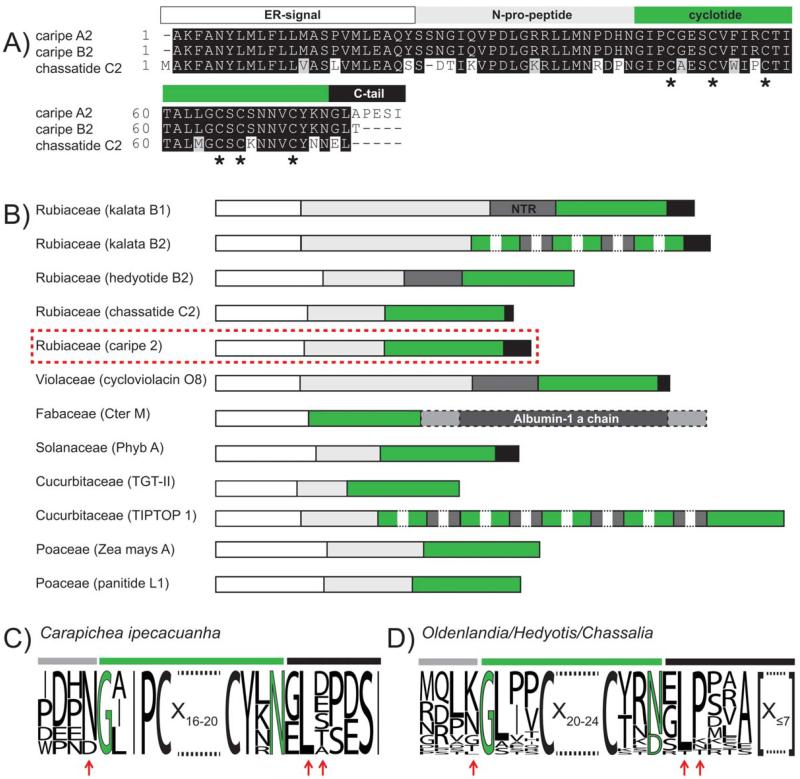
FIGURE 3. Comparison of novel Carapichea and known cyclotide precursors.
(A) A boxshade alignment of the precursors caripe A2 (plantlet, GenBank ID: KC807202), caripe B2 (root, Gen-Bank ID: KC811328), and their closest homolog precursor sequence (according to BLASTp analysis), chassatide C2 (UniProtKB ID: I0B6F2) is shown. Based on this comparison, both Carapichea sequences lack only the N-terminal methionine residue. Conserved cysteine residues are indicated with asterisks at the bottom of the alignment. (B) An overview of the architecture of cyclotide precursor proteins among different plant families is shown. Typical cyclotide precursor domains are highlighted with colors, i.e., ER signal (white), N-terminal pro peptide (light-grey), N-terminal repeats (NTR, dark-grey), mature cyclotide or knottin sequence (TGT-II and TIPTOP 1) (green), C-terminal tail sequence (black), albumin-1 a-chain (dark-grey, dashed box), and linking peptides (light-grey, dashed box). For comparison of processing residues, a sequence alignment in the form of a sequence logo for nine Carapichea ipecacuanha precursors (C) and for 25 Rubiaceae precursors (D), including those from Oldenlandia, Hedyotis, and Chassalia spp. is shown. Residues near the N- and C-terminal processing sites that show intra- or inter-species homology have been indicated by red arrows. Numbers indicate the residue length of amino acids of mature cyclotides or C-tail residues, respectively, that are not displayed for illustration purpose.