Significance
The first cells to have emerged on the early Earth lacked enzymes, and therefore probably relied on spontaneous reactions to copy their genetic material, commonly thought to have been RNA. Here we study a close chemical relative of RNA, known as 3′-phosphoramidate-DNA, in an effort to learn general lessons about nonenzymatic replication. We show that short regions of this polymer can be copied, but that the error rate is unacceptably high. We then show that a single atom change in one of the four nucleotides can restore good copying accuracy. These advances may help to achieve chemical copying of RNA, and also suggest that 3′-phosphoramidate-DNA may be a suitable genetic polymer for use in the construction of simple artificial cells.
Keywords: origin of life, nonenzymatic primer extension, artificial genetic systems, nucleotide modifications, mismatch
Abstract
Recent advances suggest that it may be possible to construct simple artificial cells from two subsystems: a self-replicating cell membrane and a self-replicating genetic polymer. Although multiple pathways for the growth and division of model protocell membranes have been characterized, no self-replicating genetic material is yet available. Nonenzymatic template-directed synthesis of RNA with activated ribonucleotide monomers has led to the copying of short RNA templates; however, these reactions are generally slow (taking days to weeks) and highly error prone. N3′-P5′–linked phosphoramidate DNA (3′-NP-DNA) is similar to RNA in its overall duplex structure, and is attractive as an alternative to RNA because the high reactivity of its corresponding monomers allows rapid and efficient copying of all four nucleobases on homopolymeric RNA and DNA templates. Here we show that both homopolymeric and mixed-sequence 3′-NP-DNA templates can be copied into complementary 3′-NP-DNA sequences. G:T and A:C wobble pairing leads to a high error rate, but the modified nucleoside 2-thiothymidine suppresses wobble pairing. We show that the 2-thiothymidine modification increases both polymerization rate and fidelity in the copying of a 3′-NP-DNA template into a complementary strand of 3′-NP-DNA. Our results suggest that 3′-NP-DNA has the potential to serve as the genetic material of artificial biological systems.
The phosphoramidate nucleic acids are of particular interest as potential genetic materials for artificial life-forms because of their potential for replication by the nonenzymatic polymerization of amino-sugar nucleotides. Because of the greater nucleophilicity of the amino group relative to the 3′-hydroxyl group of ribo- and deoxyribo-nucleotides, amino-sugar nucleotides exhibit more rapid spontaneous polymerization. Obviating the requirement for a polymerase greatly simplifies the task of creating and assembling the components of an artificial cell, and thus of constructing simple living systems from inanimate materials. We and others have therefore explored the synthesis of a variety of phosphoramidate-linked nucleic acids, their corresponding amino-sugar monomers, and the characterization of nonenzymatic template-directed primer extension reactions in these systems (1–11). Among these systems, we have examined 2′-amino versions of the acyclic glycerol nucleic acid (5), 2′-amino-2′,3′-dideoxyribonucleic acid (4, 6), and 3′-amino-2′,3′-dideoxyribonucleic acid (7).
The structural simplicity of the acyclic sugar-phosphate nucleic acid backbones has made them attractive targets for study. Indeed, an acyclic nucleotide consisting of a glycerol-phosphate backbone linked to a formylated nucleobase (12) was among the first of such nucleic acids to be chemically synthesized, but incorporation of this nucleotide into oligomers caused a severe loss of duplex stability. Much later, the glycerol nucleic acids, in which a glycerophosphate backbone is directly linked to the nucleobase via the 3′-carbon, were synthesized by Meggers and coworkers (13), and shown to form an antiparallel, Watson–Crick double-stranded helix of remarkable thermal stability, despite the added entropic cost of duplex formation. We subsequently studied the corresponding phosphoramidate polymer, based on 2′-amino substituted derivative of the glycerol monomers. However, activation of the 1′-phosphate of these monomers led to very rapid cyclization and the accumulation of inert 1′-2′-cyclic phosphates (5). Indeed, we were only able to demonstrate nonenzymatic template copying after the synthesis of activated dinucleotides. In an effort to avoid the problems associated with monomer cyclization, we turned to the 2′-amino-2′,3′-dideoxyribonucleotides, which polymerize into 2′-5′ linked phosphoramidate DNA (6). These monomers do not cyclize, as the cyclic sugar keeps the amine nucleophile away from the activated phosphate electrophile. Moreover, activation of the phosphate with an imidazole leaving group generated highly reactive monomers that engaged in rapid primer extension on DNA, RNA, and even 2′-5′ linked DNA templates. Because this template-copying reaction proceeds independently of divalent cations, which precipitate fatty acids, we were able to demonstrate template-copying inside model protocell membranes (4). Although this was an important conceptual advance toward the synthesis of a complete protocell, subsequent studies of the 2′-5′ NP-DNA system revealed several limitations. In particular, copying of A and T templates was very slow; replacement of A with 2,6 diaminopurine and of T with 5-propynyl-T resulted in faster template copying, but at the apparent cost of decreased fidelity (6).
An attractive alternative to 2′-5′ linked phosphoramidate DNA is the more RNA-like 3′-5′ linked NP-DNA. Structural studies show that this polymer forms a duplex structure that is very similar to that of RNA (14). Extensive studies by Richert and coworkers of the extension of primers ending in a 3′-amino nucleotide show that the template-directed primer extension reaction can be very fast, and can occur with relatively high fidelity on both RNA and DNA templates (8, 10). We have recently shown that short homopolymeric RNA and DNA templates can be very rapidly copied by nonenzymatic primer extension in the presence of 2-methylimidazole-activated 3′-amino nucleotides (7). Here we show that these monomers can also copy 3′-NP-DNA templates, in a true self-copying process. We also examine the fidelity of copying of mixed-sequence templates and show that wobble base pairing is a major problem, but that this problem can be largely remedied by replacing T with 2-thiothymidine (2-thio-T).
Results
3′-NP-DNA Monomers Can Copy Homopolymeric 3′-NP-DNA Templates.
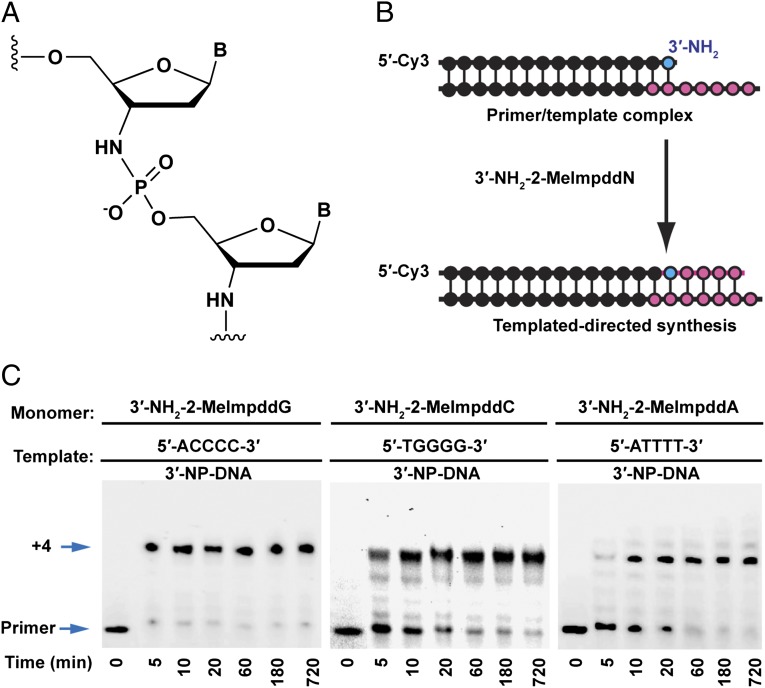
We have recently used activated 3′-amino-2′,3′-dideoxynucleotide monomers in the nonenzymatic copying of short homopolymeric DNA, RNA, and locked nucleic acid (LNA) templates (7). The copying of such templates with 2-methylimidazole-activated monomers was rapid and accurate, with superior copying on RNA and LNA templates as opposed to DNA templates. These observations prompted us to investigate the copying of 3′-NP-DNA templates into complementary 3′-NP-DNA, using 5′-phosphor-(2-methyl)imidazolides of the 3′-amino-2′,3′-dideoxynucleotides as activated monomers. Due to the relatively low efficiency of solid-phase synthesis of 3′-NP-DNA, we used chimeric oligonucleotides in which only the 3′-end of the primer and the 5′-end of the template were composed of 3′-NP-DNA (Fig. 1 A and B). A DNA primer bearing a single 3′-amino-2′,3′-dideoxynucleotide at its 3′ terminus was generated by solid-phase synthesis using 5′-phosphoramidites. This primer was annealed to an oligonucleotide with a primer-binding DNA region and a seven-nucleotide 3′-NP-DNA region, including a four-nucleotide template region. To minimize the structural discontinuity between the DNA duplex and the NP-DNA duplex, the two nucleotides in the template oligonucleotide that pair with the last two nucleotides of the primer were also 3′-NP-DNA. An additional 5′-terminal nucleotide was added to the template to provide favorable stacking interactions. Upon the addition of 2-methylimidazole-activated monomers (3′-NH2-2-MeImpddNs) to the primer-template complex, the primer was extended by the spontaneous template-directed synthesis of 3′-NP-DNA. The reaction products were analyzed by PAGE, and their identity was confirmed by liquid chromatography–mass spectrometry (LC-MS).
Fig. 1.
Nonenzymatic primer extension reactions on homopolymeric templates. (A) Structure of an internucleotide linkage in N3′-P5′–linked phosphoramidate DNA (3′-NP-DNA). (B) General primer extension reaction scheme showing a 5′-Cy3–labeled 3′-amino–terminated DNA primer annealed to a complementary chimeric DNA/3′-NP-DNA template. The 3′-NH2-2-MeImpddN monomers extend the primer by four nucleotides on the complementary template forming a chimeric DNA/3′-NP-DNA product. Red lines indicate phosphoramidate bonds. (C) PAGE analysis of primer extension products on indicated 3′-NP-DNA templates. Primer extension reactions contained 0.1 µM Cy3-labeled-3′-amino–terminated DNA primer, 0.5 µM template, 100 mM Mes-CAPS-Hepes, pH 7.5, and 100 mM 1-(2-hydroxyethyl)imidazole (HEI). The reactions were initiated by addition of 5.0 mM 3′-NH2-2-MeImpddG, 5.0 mM 3′-NH2-2-MeImpddC, and 10.0 mM 3′-NH2-2-MeImpddA, respectively. Arrows indicate the primer and the full-length product (+4).
Activated 3′-amino-2′,3′-dideoxynucleotides are just as effective in copying 3′-NP-DNA templates as they are in copying DNA and RNA templates in nonenzymatic primer extension reactions. Of the four 2-methylimidazolide monomers, 3′-NH2-2-MeImpddG was the most reactive, reaching >92% completion within 5 min while copying a C4 3′-NP-DNA template (Fig. 1C and SI Appendix, Fig. S1). The C, T, and A monomers also resulted in efficient copying of their complementary homopolymeric 3′-NP-DNA templates, achieving ≥80% completion in less than an hour (Figs. 1C and 2). The efficiency of extension on 3′-NP-DNA templates was comparable to that on RNA templates, and was higher than that on DNA templates (SI Appendix, Figs. S2–S4). As previously observed (7), in some cases the kinetics of primer extension are unusual in that the reaction appears to progress from unreacted primer to fully extended primer, with the expected intermediates present at low or undetectable levels. Such kinetics could reflect slow initiation of primer extension, followed by rapid elongation, possibly due to a structural discontinuity between the DNA/DNA primer/template region, and the 3′-NP-DNA portion of the template. As an alternate, monomers may assemble into oligomers on the 3′-NP-DNA template, but only slowly ligate to the primer. The fact that 3′-NP-DNA can serve as a template for its own synthesis inspired us to look more deeply into the properties of this self-copying reaction.
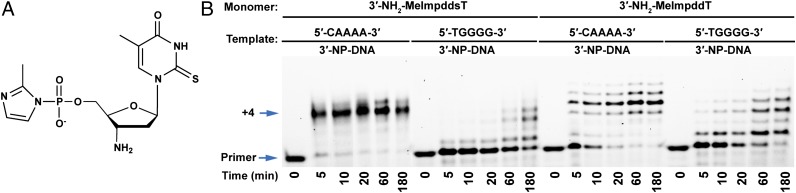
Fig. 2.
Comparison of nonenzymatic primer extension reactions on homopolymeric A4 and G4 templates using T and 2-thio-T monomers. (A) Chemical structure of monomer 3′-NH2-2-MeImpddsT. (B) PAGE analysis of primer extension products on indicated templates. Primer extension reactions were carried out as in Fig. 1. The reaction was initiated by addition of 10.0 mM 3′-NH2-2-MeImpddsT or 3′-NH2-2-MeImpddT. Arrows indicate primer and full-length product. Primer extension is faster using 2-thio-T vs. T on the A4 template, but slower on the G4 template.
2-thio-T Replacement of T Enhances Copying of Correct Reactions and Suppresses Incorrect Copying Reactions.
Our previous observation of polymerization of 3′-NH2-2-MeImpddT on a G4 RNA template suggested that the formation of G:T wobble base pairs would be a significant problem in the 3′-NP-DNA system (7). Thermodynamic data indicate that 2-thio-U (sU) forms a stronger base pair with A than with standard U by ∼1 kcal/mol, and the G:sU wobble pair is slightly destabilized relative to G:U within RNA duplexes (15). Similarly, in DNA duplexes, the A:sT base pair is stabilized relative to A:T, and the G: sT wobble pair is significantly destabilized relative to G:T (16, 17). We therefore asked whether activated 2-thio-T monomer (3′-NH2-2-MeImpddsT, a compound not previously used in primer extension reactions) might allow more rapid and efficient copying of A residues in 3′-NP-DNA templates as a result of the higher stability of the sT:A base pair, and reducing misincorporation due to G:T wobble pairing in nonenzymatic template-directed primer extension reactions. It is remarkable that the copying of the A4 3′-NP-DNA template (Fig. 2) with the 2-thio-T monomer 3′-NH2-2-MeImpddsT was almost complete in 5 min, compared with 1 h using 3′-NH2-2-MeImpddT. Similar increases in the rate of primer extension were seen at shorter times and with lower monomer concentrations (SI Appendix, Figs. S5–S7). We observed extensive copying of a G4 3′-NP-DNA template using the normal T monomer 3′-NH2-2-MeImpddT. It was gratifying that this incorrect copying reaction was almost completely suppressed when we used the 2-thio monomer 3′-NH2-2-MeImpddsT.
2-Thio-dT/U Enhances Copying Fidelity When Included in Monomer or Template Contexts.
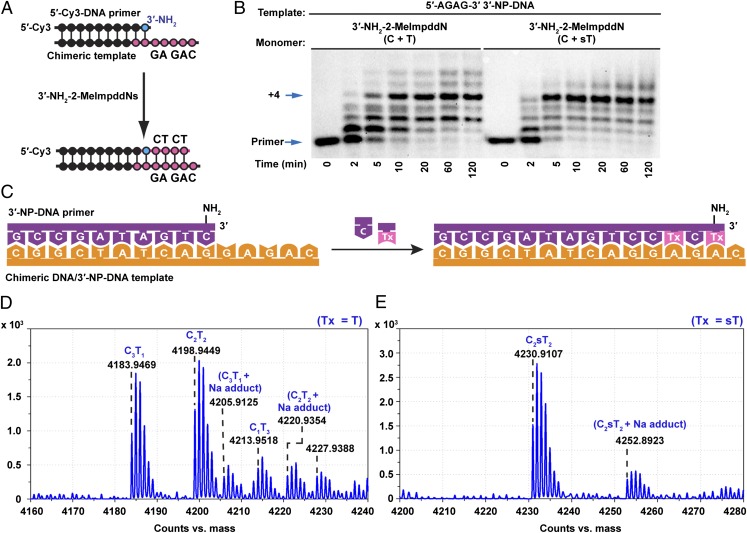
Encouraged by the excellent rates and yields for copying homopolymeric templates, we proceeded to examine the copying of mixed-sequence templates using 3′-amino monomers. We first focused on the extension of a primer over a 5′-AGAG-3′ sequence on a 3′-NP-DNA template (Fig. 3). An initial time course using a 5′-Cy3 labeled primer and PAGE to separate the primer from extended products suggested that the four-nucleotide template region was rapidly and efficiently copied (about 70% + 4 product after 20 min), using a mixture of 5.0 mM 3′-NH2-2-MeImpddC and 5.0 mM 3′-NH2-2-MeImpddT monomers. However, in light of the expected high frequency of G:T mismatches, we conducted a larger scale reaction using an unlabeled 3′-NP-DNA primer for MS analysis. The correct product would consist of the primer plus 2 C and 2 T residues; however, this C2T2 product comprised only 48% of the primer + 4 nucleotide (N+4) products (Fig. 3D and SI Appendix, Fig. S8). We also detected 6% of a C1T3 product, consisting of the primer extended by 1 C and 3 T residues caused by a G:T mismatch. We were surprised to observe 42% of a C3T1 product caused by an A:C mismatch, as well as 5% of a C4 product resulting from two A:C mismatches during copying of the AGAG 3′-NP-DNA template. It is possible that the relatively slow copying of template As by a T monomer allowed time for the insertion of Cs by wobble pairing. In an effort to reduce the A:C mismatched products and improve fidelity, we raised the concentration of the T monomer to 20.0 mM and kept the C monomer at 5.0 mM. Under these conditions, we found that no A:C mismatched C4 product could be observed within the limit of detection of the instrument, and that the A:C mismatched product C3T1 was indeed reduced to 26% (SI Appendix, Fig. S8), but the C1T3 product caused by a G:T mismatch increased to 16%. Overall the correct C2T2 product was 58% of the +4 material, only a slight enhancement of fidelity as a result of changing the monomer C/T ratio. These results indicate that the fidelity of template copying with the canonical C and T monomers is unacceptably low in nonenzymatic copying of 3′-NP-DNA templates.
Fig. 3.
Faster and more accurate copying of a 3′-NP-DNA template using 2-thio-T in place of T monomer. (A) Scheme for self-copying of a 3′-NP-DNA template using C and T (or 2-thio-T) monomers. The primer is a 5′-Cy3-DNA-3′-NH2 and the template is a chimeric DNA/3′-NP-DNA (underlined) 5′-CAGAGGACTATCGGC-3′. (B) PAGE analysis of primer extension products. Primer extension reactions were carried out as in Fig. 1. The reaction was initiated by addition of 5.0 mM 3′-NH2-2-MeImpddC and 5.0 mM 3′-NH2-2-MeImpddT/sT as indicated. Arrows indicate primer and full-length product. (C) Scheme for self-copying AGAG 3′-NP-DNA template with an entirely 3′-NP-DNA primer. (D) High-resolution MS analysis of the full-length extension products from a reaction initiated by the addition of 5.0 mM 3′-NH2-2-MeImpddC and 5.0 mM 3′-NH2-2-MeImpddT. There are three major isomeric families for three different 14-mer full-length N+4 products, corresponding to C3T1, C2T2, and C1T3 respectively. (E) High-resolution MS analysis of the full-length extension products from a reaction initiated by the addition of 5.0 mM 3′-NH2-2-MeImpddC and 5.0 mM 3′-NH2-2-MeImpddsT. The major peak corresponds to the 14-mer full-length N+4 product C2sT2.
In an effort to improve the fidelity of copying of the 5′-AGAG template, we used the activated 2-thio-T monomer 3′-NH2-2-MeImpddsT. Indeed, the copying of the 5′-AGAG 3′-NP-DNA template was significantly faster and more efficient (about 90% +4 product after 10 min) using 5.0 mM 3′-NH2-2-MeImpddC and 5.0 mM of the 2-thio monomer 3′-NH2-2-MeImpddsT when compared to the normal T monomer (Fig. 3B). Most importantly, MS analysis revealed that the fidelity was enhanced dramatically with the 2-thio-T monomer. We observed mainly the correct C2sT2 product (82%) by MS analysis of the full-length N+4 products (Fig. 3E and SI Appendix, Fig. S8), corresponding to an average fidelity of over 95% per position. The C3sT1 product resulting from an A:C mismatch was reduced to 12%, compared with 42% when the T monomer was used. We did not observe any C4 product within the limit of detection of the instrument. To further confirm that the product was the expected primer–CsTCsT product (and not an isomeric mismatched product), we sequenced the 3′-NP-DNA product by partial acid hydrolysis of the N3′-P5′ phosphoramidate bonds (18) followed by LC-MS analysis of the resulting sequence ladder. The sequencing data confirmed that the product was the expected primer–CsTCsT product (SI Appendix, Table S1), showing directly that the two monomers were added to the primer faithfully while copying the 3′-NP-DNA template. A similarly high proportion (85%) of correct N+4 products was also observed when a mixture of 5.0 mM 3′-NH2-2-MeImpddsT and 5.0 mM 3′-NH2-2-MeImpddC was used to copy a 5′-AGAG RNA template, and the proportion of correct product was greater than 95% when copying a DNA template under the same conditions (SI Appendix, Fig. S8).
The enhanced fidelity that we observed with the 2-thio-T monomer led us to examine the copying of templates containing T and 2-thio-T residues. We first examined the fidelity of nonenzymatic copying of a 5′-TCTC 3′-NP-DNA template using an optimized monomer ratio of 1.0 mM 3′-NH2-2-MeImpddG and 10.0 mM 3′-NH2-2-MeImpddA. LC-MS analysis of the products of primer extension showed that the expected A2G2 product accounted for 43% of the N+4 product (SI Appendix, Fig. S9), while the mismatched products A4, A3G1, A1G3, and G4 accounted for 1%, 26%, 27%, and 2%, respectively. Similarly poor fidelity was also observed using DNA and RNA templates with the same sequences (SI Appendix, Fig. S9). Unfortunately, synthetic problems in the synthesis of the 3′-amino-2-thiothymidine-5′-O-phosphoramidite (SI Appendix) have so far made it impossible to replace T with 2-thio-T in 3′-NP-DNA oligonucleotides. We therefore compared the fidelity of 3′-NP-DNA synthesis in the copying of RNA templates containing either U or 2-thio-U.
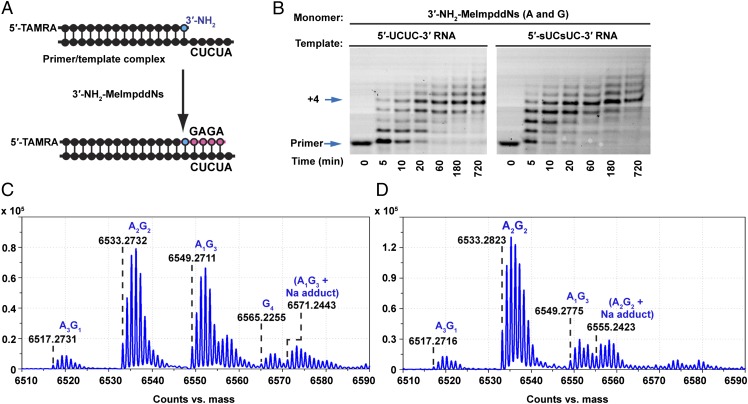
We compared primer extension on 5′-UCUC and 5′-sUCsUC templates, using a mixture of 5.0 mM 3′-NH2-2-MeImpddG and 10.0 mM 3′-NH2-2-MeImpddA monomers (Fig. 4). We observed 90% full-length product at 1 h on the 2-thio-U–containing template, compared with 84% full-length product on the native RNA template over the course of 3 h (Fig. 4B). As expected, LC-MS studies of the primer +4 product from the normal RNA 5′-UCUC template revealed four major isomeric families of full-length products, corresponding to the desired A2G2, as well as A3G1, A1G3, and G4 (Fig. 4C). The correct A2G2 product accounted for 45% of the full-length N+4 products (SI Appendix, Fig. S9). A larger portion of N+4 products in the copying reaction resulted from G:T mismatches, with 41% A1G3 and 8% G4, consistent with the high levels of G:T wobble pairing previously observed in nonenzymatic primer extension experiments (7, 19). We also detected 5% A3G1, presumably due to A:C mismatch pairing.
Fig. 4.
Enhanced copying fidelity of an RNA template containing 2-thioU in place of U. (A) Scheme for nonenzymatic primer extension on UCUC (or sUCsUC) RNA templates. Red segment indicates newly formed phosphoramidate bonds. (B) PAGE analysis of primer extension products. Primer extension reactions were carried out as in Fig. 1. The reaction was initiated by addition of 5.0 mM 3′-NH2-2-MeImpddG and 10.0 mM 3′-NH2-2-MeImpddA. Arrows indicate primer and full-length product. (C) High-resolution MS analysis of the full-length extension products from a reaction of 100 pmol primer extended on a UCUC RNA template for 12 h followed by ethanol precipitation. There are four distinct major families of isotopic peaks for four different 14-mer full-length N+4 products, corresponding to A3G1, A2G2, A1G3, and G4, respectively. (D) High-resolution MS analysis of the full-length extension products from copying a sUCsUC RNA template. The major peak corresponds to the N+4 full-length product A2G2.
The fidelity of template copying improved dramatically on the 2-thio-U containing RNA template (Fig. 4D). When a mixture of 5.0 mM 3′-NH2-2-MeImpddG and 15.0 mM 3′-NH2-2-MeImpddA was used to copy a 5′-sUCsUC RNA template, the proportion of correct full-length N+4 products was 92% (SI Appendix, Fig. S9), corresponding to a per-site average fidelity of ∼98%. To confirm that the major product was the Watson–Crick GAGA product, we sequenced the 3′-NP-DNA products as previously described. LC-MS sequencing confirmed that the A2G2 product had the correct primer-GAGA sequence (SI Appendix, Table S2). The similarity of 3′-NP-DNA and RNA templates leads us to expect that similarly accurate copying would be observed with 3′-NP-DNA templates containing 2-thio-T in place of T.
The replication of arbitrary sequences of a nucleic acid requires accurate copying of mixed-sequence templates in the presence of all four different monomers (i.e., A, C, G, and T/sT). Therefore, we used first three, and then four different monomers in copying reactions to assess fidelity in this more complex situation. When we added 1.25 mM of the G monomer to 5.0 mM of the C and T monomers to copy the 5′-AGAG 3′-NP-DNA template, only 38% of the full-length N+4 product had the expected C2T2 composition. The majority of N+4 products in the copying reaction resulted from G:T mismatches, with 49% C1T3 and 7% T4. We also detected 7% C3T1 caused by an A:C mismatch (SI Appendix, Fig. S8). The fidelity was enhanced significantly when the T monomer was replaced with sT. The expected C2sT2 product accounted for 79% of the full-length N+4 products, and the G:T mismatched product C1sT3 was reduced to only 9%, although the A:C mismatched products increased slightly (2% C4 and 11% C3sT1) (SI Appendix, Fig. S8). The product mixture was even more complicated when we used 5.0 mM A, C, and T, and 1.25 mM G monomers. The expected C2T2 product accounted only for 25% of the full-length N+4 products (SI Appendix, Fig. S8). We observed largely G:T mismatch products (56% C1T3 and 8% T4), and 6% C3T1 and 1% C4 resulting from A:C mismatch pairing as well as 4% A1C2T1. As above, the fidelity was greatly enhanced when we replaced T with sT under the same conditions (SI Appendix, Fig. S8). We observed 74% of the expected C2sT2 product, and G:T mismatched products were reduced to only 9% of C1sT3; a small increase of A:C mismatched products was also observed (3% C4 and 15% C3sT1). These results indicate that even in the presence of all four monomers, reasonable fidelity can be achieved in the nonenzymatic copying of mixed templates, as long as T is replaced by 2-thio-T.
Discussion
In addition to suppressing G:T mismatches, we were initially surprised to find that 2-thio-T also reduced A:C mismatches. We attribute the suppression of A:C mismatch products to stronger A:sT base pairing, so that 2-thio-T outcompetes C for binding to A. This is consistent with our observations on the copying of homopolymer templates, where under the same conditions the reactivity of the 2-thio-T monomer on A4 templates was much faster than that of the normal T monomer, and was similar to that of the C monomer on G4 templates (SI Appendix, Figs. S5–S7). Replacement of T with 2-thio-T also enhanced both rate and yield in replicating the 5′-AGAG 3′-NP-DNA template. Thus, the increased fidelity of nonenzymatic copying of 3′-NP-DNA with 2-thio-T is most likely due to the combination of decreased G:sT wobble pairing, and stronger A:sT Watson–Crick pairing, which decreases the opportunity for A:C mismatches to form.
Our results suggest that the 2-thio modification of U (or T) might have played a role in primordial RNA replication processes. It is interesting that 2-thio-U and 2-seleno-U (20) are found in the third (wobble) position of the anticodons of specific tRNAs, where they may contribute to enhanced affinity and specificity of the codon–anticodon interaction (21, 22). The modulation of base-pair affinity and specificity by simple chemical modifications may have been particularly important during the emergence of the RNA world, before the evolution of polymerase-catalyzed replication. It is an intriguing speculation that the continued use of these modifications in extant biochemistry may be a relic of the chemical origins of life (23).
The fast and accurate nonenzymatic copying of short mixed-sequence 3′-NP-DNA templates suggests that 3′-NP-DNA may be a suitable genetic polymer for use in the construction of artificial cellular systems. Assessing the genomic potential of 3′-NP-DNA will require studies of the copying of longer 3′-NP-DNA templates with varied sequences. Such studies are currently limited by the difficulty of solid-phase synthesis of 3′-NP-DNA, which is expensive and inefficient. However, the nonenzymatic synthesis of 3′-NP-DNA by copying RNA or LNA templates may provide an alternative route to the synthesis of 3′-NP-DNA oligonucleotides if this can be carried out with sufficient fidelity and on an appropriate scale. A potential barrier to 3′-NP-DNA replication is the difficulty of strand separation due to the high Tm of duplex 3′-NP-DNA (24–26), and the additional duplex stabilization expected from 2-thio-T substitution. This problem may be ameliorated by moderate concentrations of formamide or urea (27), or alternatively, a heterogeneous backbone of N3′-P5′ and N2′-P5′ linkages could be used (6). The acid lability of 3′-NP-DNA would constrain protocells based on this genetic polymer to neutral or moderately alkaline environments. If complete cycles of 3′-NP-DNA replication can be achieved, it may be possible to assemble simple protocells (4, 28) within which genomic information resides in a nonbiological polymer.
Materials and Methods
Synthesis of Monomers and 3′-NP-DNA Primer and Templates.
Here, 3′-NH2-2-MeImpddNs (A, C, G, and T) were synthesized as previously described (7). The synthesis of 3′-NH2-2-MeImpddsT is described in SI Appendix. The 3′-NP-DNA primers and templates (29, 30) were synthesized at Geron Corp., and were purified by reverse-phase HPLC (Agilent 1100 series LC) on a 50 × 4.6-mm C18 column (XTerra) at 25 °C.
Nonenzymatic Primer Extension Reactions.
Template copying reactions contained 0.1-μM 5′-Cy3/carboxytetramethylrhodamine (TAMRA)-labeled 3′-amino–terminated primer, 0.5-μM template oligonucleotide, 150 mM NaCl, 100 mM HEI, 100 mM 2-(N-morpholino)ethanesulfonic acid/N-cyclohexyl-3-aminopropanesulfonic acid/4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (MES-CAPS-Hepes), pH 7.5, and 3′-NH2-2-MeImpddNs at indicated concentrations. Reactions were initiated by addition of activated monomers and incubated at 4 °C. Aliquots were removed and stopped at indicated time points by addition of three volumes of formamide and heating to 95 °C for 5 min, followed immediately by ethanol precipitation on dry ice. Stopped reactions were resuspended in 8.0 M urea and heated to 95 °C for 5 min. Samples were analyzed by electrophoresis on 7.0 M urea, 17% (wt/vol) polyacrylamide sequencing gels. Reaction products were visualized by fluorescence imaging on a Typhoon 9410 PhosphorImager using the Cy3 fluorophore filter set. Product quantification and analysis was performed using ImageQuant TL software (GE Healthcare Life Sciences).
LC-MS Studies of Fidelity in Nonenzymatic Primer Extension Reactions.
Primer extension products analyzed by LC-MS were prepared by extending 40–100 pmol of 3′-amino–terminated DNA primer at 4 °C for 12 h on complementary templates in similar conditions to those previously described. Crude samples were injected for LC-MS analysis after ethanol precipitation. LC-MS measurements were performed using an Agilent 6520 Q-TOF mass analyzer and 1200 series HPLC with a Waters XBridge C18 column (3.5 μm, 1 × 100 mm). Mobile phase A was aqueous 200 mM hexafluoroisopropanol and 3 mM triethylamine at pH 7.0, and mobile phase B was methanol. The HPLC method for 35 μL of a 2.5 μM solution was a linear increase of 5–20% B over 30 min at 0.1 mL/min, with the column heated to 60 °C. Sample elution was monitored by absorbance at 260 nm and the eluate was passed directly to an electrospray ionization source with 325 °C drying nitrogen gas flowing at 8.0 L/min, a nebulizer pressure of 30 pounds per square inch gauge and a capillary voltage of 3500 V. Agilent MassHunter Qualitative Analysis software was used to analyze Q-TOF–derived MS data. The relative quantities of different product species were quantified for fidelity measurements by integrating the extracted ion current peak for a 0.3 mass-to-charge ratio (m/z) range centered on the −3 charge state of the 13C2 isotopes (for the 3′-NP-DNA primer) or a 0.26 m/z range centered on the −4 charge state of the 13C3 isotopes (for the TAMRA-labeled DNA primer). The narrow ranges were used to minimize overlap between peaks; however, the relative values obtained were very similar when larger ranges were used.
Acidic Hydrolysis of 3′-NP-DNA for LC-MS Sequencing.
The 3′-NP-DNA primer extension products were prepared for sequence analysis from a reaction of 200 pmol 3′-amino–terminated primer extended on mixed templates for 12 h followed by ethanol precipitation. To the crude 3′-NP-DNA product (8 μL, 25 μM) in a 0.2 mL Eppendorf tube was added 2 μL 25% acetic acid. The solution was vortexed, and heated in a water bath at 40 °C for 30 min. The resultant partially degraded products were subjected to LC-MS analysis after ethanol precipitation.
Supplementary Material
Acknowledgments
We thank members of our laboratory for helpful discussions and comments on the manuscript. This research was funded in part by Grant CHE-0809413 from the National Science Foundation. J.W.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312329110/-/DCSupplemental.
References
- 1.Lohrmann R, Orgel LE. Template-directed synthesis of high molecular weight polynucleotide analogues. Nature. 1976;261(5558):342–344. doi: 10.1038/261342a0. [DOI] [PubMed] [Google Scholar]
- 2.Zielinski WS, Orgel LE. Oligomerization of activated derivatives of 3′-amino-3′-deoxyguanosine on poly(C) and poly(dC) templates. Nucleic Acids Res. 1985;13(7):2469–2484. doi: 10.1093/nar/13.7.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tohidi M, Zielinski WS, Chen CH, Orgel LE. Oligomerization of 3′-amino-3’deoxyguanosine-5’phosphorimidazolidate on a d(CpCpCpCpC) template. J Mol Evol. 1987;25:97–99. doi: 10.1007/BF02101750. [DOI] [PubMed] [Google Scholar]
- 4.Mansy SS, et al. Template-directed synthesis of a genetic polymer in a model protocell. Nature. 2008;454(7200):122–125. doi: 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen JJ, Cai X, Szostak JW. N2′—>p3′ phosphoramidate glycerol nucleic acid as a potential alternative genetic system. J Am Chem Soc. 2009;131(6):2119–2121. doi: 10.1021/ja809069b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrum JP, Ricardo A, Krishnamurthy M, Blain JC, Szostak JW. Efficient and rapid template-directed nucleic acid copying using 2′-amino-2′,3′-dideoxyribonucleoside-5′-phosphorimidazolide monomers. J Am Chem Soc. 2009;131(40):14560–14570. doi: 10.1021/ja906557v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Zhang N, Blain JC, Szostak JW. Synthesis of N3′-P5′-linked phosphoramidate DNA by nonenzymatic template-directed primer extension. J Am Chem Soc. 2013;135(2):924–932. doi: 10.1021/ja311164j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Röthlingshöfer M, et al. Chemical primer extension in seconds. Angew Chem Int Ed Engl. 2008;47(32):6065–6068. doi: 10.1002/anie.200801260. [DOI] [PubMed] [Google Scholar]
- 9.Eisenhuth R, Richert C. Convenient syntheses of 3′-amino-2′,3′-dideoxynucleosides, their 5′-monophosphates, and 3′-aminoterminal oligodeoxynucleotide primers. J Org Chem. 2009;74(1):26–37. doi: 10.1021/jo8018889. [DOI] [PubMed] [Google Scholar]
- 10.Kaiser A, Spies S, Lommel T, Richert C. Template-directed synthesis in 3′- and 5′-direction with reversible termination. Angew Chem Int Ed Engl. 2012;51(33):8299–8303. doi: 10.1002/anie.201203859. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Guntha S, Ferencic M, Krishnamurthy R, Eschenmoser A. Base-pairing systems related to TNA: Alpha-threofuranosyl oligonucleotides containing phosphoramidate linkages. Org Lett. 2002;4(8):1279–1282. doi: 10.1021/ol020015x. [DOI] [PubMed] [Google Scholar]
- 12.Schneider KC, Benner SA. Oligonucleotides containing flexible nucleoside analogues. J Am Chem Soc. 1990;112(1):453–455. [Google Scholar]
- 13.Zhang L, Peritz A, Meggers E. A simple glycol nucleic acid. J Am Chem Soc. 2005;127(12):4174–4175. doi: 10.1021/ja042564z. [DOI] [PubMed] [Google Scholar]
- 14.Tereshko V, Gryaznov S, Egli M. Consequences of replacing the DNA 3'-oxygen by an amino group: High-resolution crystal structure of a fully modified N3'-> P5' phosphoramidate DNA dodecamer duplex. J Am Chem Soc. 1998;120(2):269–283. [Google Scholar]
- 15.Testa SM, Disney MD, Turner DH, Kierzek R. Thermodynamics of RNA-RNA duplexes with 2- or 4-thiouridines: Implications for antisense design and targeting a group I intron. Biochemistry. 1999;38(50):16655–16662. doi: 10.1021/bi991187d. [DOI] [PubMed] [Google Scholar]
- 16.Lezius AG. Synthesis and characterisation of a copolymer consisting of alternating deoxyadenosine- and 2-thiodeoxythymidine nucleotides. Eur J Biochem. 1970;14(1):154–160. doi: 10.1111/j.1432-1033.1970.tb00273.x. [DOI] [PubMed] [Google Scholar]
- 17.Sintim HO, Kool ET. Enhanced base pairing and replication efficiency of thiothymidines, expanded-size variants of thymidine. J Am Chem Soc. 2006;128(2):396–397. doi: 10.1021/ja0562447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfe JL, Kawate T, Belenky A, Stanton V., Jr Synthesis and polymerase incorporation of 5′-amino-2′,5′-dideoxy-5′-N-triphosphate nucleotides. Nucleic Acids Res. 2002;30(17):3739–3747. doi: 10.1093/nar/gkf502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leu K, Obermayer B, Rajamani S, Gerland U, Chen IA. The prebiotic evolutionary advantage of transferring genetic information from RNA to DNA. Nucleic Acids Res. 2011;39(18):8135–8147. doi: 10.1093/nar/gkr525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan AE, Sheng J, Zhang W, Huang Z. High fidelity of base pairing by 2-selenothymidine in DNA. J Am Chem Soc. 2010;132(7):2120–2121. doi: 10.1021/ja909330m. [DOI] [PubMed] [Google Scholar]
- 21.Caton-Williams J, Huang Z. Biochemistry of selenium-derivatized naturally occurring and unnatural nucleic acids. Chem Biodivers. 2008;5(3):396–407. doi: 10.1002/cbdv.200890040. [DOI] [PubMed] [Google Scholar]
- 22.Ajitkumar P, Cherayil JD. Thionucleosides in transfer ribonucleic acid: Diversity, structure, biosynthesis, and function. Microbiol Rev. 1988;52(1):103–113. doi: 10.1128/mr.52.1.103-113.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szostak JW. The eightfold path to non-enzymatic RNA replication. J Syst Chem. 2012;3:2. [Google Scholar]
- 24.Gryaznov S, Chen JK. Oligodeoxyribonucleotide N3′->P5′ phosphoramidates: Synthesis and hybridization properties. J Am Chem Soc. 1994;116(7):3143–3144. [Google Scholar]
- 25.Gryaznov SM, et al. Oligonucleotide N3′—>P5′ phosphoramidates. Proc Natl Acad Sci USA. 1995;92(13):5798–5802. doi: 10.1073/pnas.92.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gryaznov SM, Letsinger RL. Synthesis and properties of oligonucleotides containing aminodeoxythymidine units. Nucleic Acids Res. 1992;20(13):3403–3409. doi: 10.1093/nar/20.13.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powner MW, Sutherland JD, Szostak JW. Chemoselective multicomponent one-pot assembly of purine precursors in water. J Am Chem Soc. 2010;132(46):16677–16688. doi: 10.1021/ja108197s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansy SS, Szostak JW. Reconstructing the emergence of cellular life through the synthesis of model protocells. Cold Spring Harb Symp Quant Biol. 2009;74:47–54. doi: 10.1101/sqb.2009.74.014. [DOI] [PubMed] [Google Scholar]
- 29.Zielinska D, Pongracz K, Gryaznov SM. A new approach to oligonucleotide N3′->P5′ phosphoramidate building blocks. Tetrahedron Lett. 2006;47:4495–4499. doi: 10.1081/ncn-200060068. [DOI] [PubMed] [Google Scholar]
- 30.Matray TJ, Gryaznov SM. Synthesis and properties of RNA analogs-oligoribonucleotide N3′—>P5′ phosphoramidates. Nucleic Acids Res. 1999;27(20):3976–3985. doi: 10.1093/nar/27.20.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.