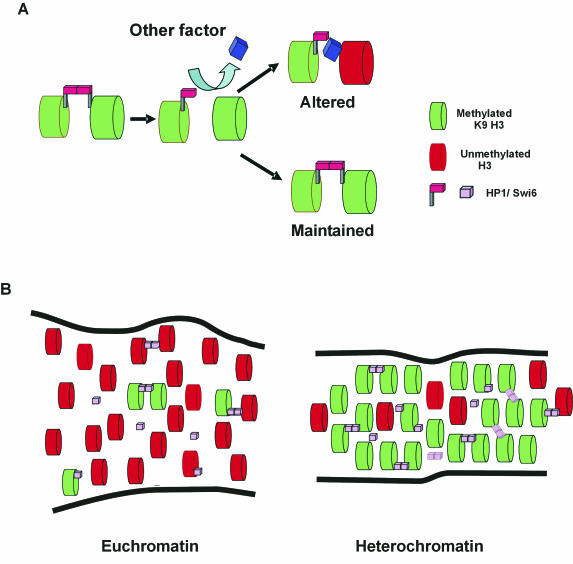
FIG. 6.
Stochastic model for HP1/Swi6 binding to chromatin. (A) HP1/Swi6 dimers cross-link adjacent nucleosomes. The dynamic nature of binding frequently creates vacant binding sites. Competition for the open binding sites determines the fate of the nucleosome. Association of HP1/Swi6 maintains the status quo. Association of a competitor results in an altered nucleosome configuration. (B) Steady-state representation of stochastic HP1/Swi6 binding to euchromatin and heterochromatin domains. HP1/Swi6 has a higher affinity for mK9-H3, and, since this compartment is enriched in mK9-H3, HP1/Swi6 has a higher probability to bind heterochromatin. Since the compaction of the nucleosome is higher in heterochromatin, the probability of establishing a cross-link to an adjacent nucleosome, either on the same fiber or a different chromatin fiber, is also higher in heterochromatin than in euchromatin. Because euchromatin is enriched in nucleosomes bearing histone H3 unmethylated on K9, the fraction of HP1 bound to unmethylated histone H3 on K9 is higher in euchromatin than in heterochromatin. These interactions are less stable than the interactions with methylated K9-H3 and do not result in heterochromatinization. The overall stability of heterochromatin is conferred by the stable methylation of core histones.