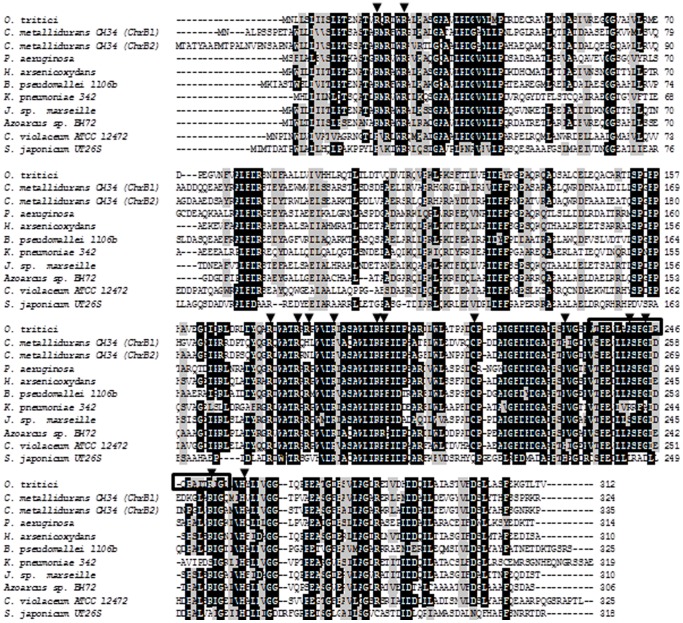
Figure 7. Alignment of ChrB homologues from different organisms.
Amino acid sequences (obtained from NCBI) were aligned via CLUSTAL W. Ochrobactrum tritici 5bvl1 (ABO70326), Pseudomonas aeruginosa (AEQ93502); Cupriavidus metallidurans CH34, ChrB1 and ChrB2 (ABF13062 and ABF10734, respectively); Herminiimonas arsenicoxydans (CAL61077); Burkholderia pseudomallei 1106b (EES21856); Klebsiella pneumoniae 342 (YP_002238121); Janthinobacterium sp. Marseille (ABR91486); Azoarcus sp. BH72 (CAL95580); Chromobacterium violaceum ATCC 12472 (AAQ58595); Sphingobium japonicum UT26S (BAI96957). Highly conserved residues in ChrB homologues (black shading), similar residues in ChrB homologues (grey shading), conserved residues chosen for mutagenesis approach (arrow) and the putative HTH motif (into the box).