| Instrument |
X33 (EMBL, DORIS ring, DESY) |
| Beam geometry |
|
2 mm×0.6 mm |
| Wavelength [Å] |
|
1.5 |
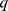 range [Å−1] range [Å−1] |
|
0.0074-0.5 |
| Exposure time [s] |
|
8×15 |
| Temperature [K] |
|
283 |
| Concentration range [mg/ml] |
|
2.2–3.8 |
|
|
0.7–1.2 |
|
Structural parameters
|
|
|
|
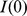 [% [% 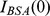 ] from P(r) ] from P(r) |
|
53±5 |
32.25±12 |
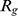
|
|
| [Å ] from P(r) |
|
31.2±1.2 |
18.8±0.1 |
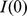 [% [% 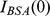 ] from Guinier ] from Guinier |
|
72.8±9 |
31.6±0.06 |
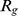
|
|
| [Å ] from Guinier |
|
29.3±2 |
17.8±2.1 |
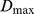 [Å] from data [Å] from data |
|
117 |
61.4 |
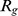
|
|
| [Å ] |
Model 1
|
30 |
17.4 |
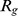
|
|
| [Å ] |
Model 2
|
33 |
|
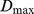 [Å] of model envelope [Å] of model envelope |
Model 1
|
112 |
61.43 |
|
Model 2
|
124 |
|
| Porod volume estimate [Å] |
|
30230 |
10950 |
| Dry volume calculated from model [Å] |
Both models
|
28900 |
14100 |
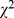 fit fit |
Model 1 |
0.75 |
1.07 |
|
Model 2 |
0.93 |
|
|
Molecular-mass determination
|
|
|
Molecular mass 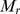 from I(0) in P(r) [kDa] from I(0) in P(r) [kDa] |
|
18±2 |
10.7±3 |
Calculated monomeric 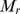 from sequence [kDa] from sequence [kDa] |
|
23.4 |
10.3 |
Calculated monomeric 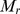 from I(0) in Guinier [kDa] from I(0) in Guinier [kDa] |
|
25±3 |
11.6 |
|
Software employed
|
|
|
| Primary data reduction |
|
PRIMUS |
| Data processing |
|
GNOM |
| Ab initio analysis |
|
DAMMIF |
| Model comparison |
|
SUPCOMB |
| Tertiary structure modelling |
|
RFR (this paper)
|
| Computation of model intensities |
|
CRYSOL |
| Three-dimensional graphics representations |
|
PyMol |
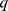 range [Å−1]
range [Å−1]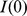 [%
[% 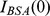 ] from P(r)
] from P(r)
 [%
[% 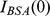 ] from Guinier
] from Guinier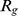
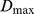 [Å] from data
[Å] from data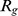
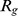
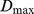 [Å] of model envelope
[Å] of model envelope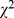 fit
fit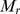 from I(0) in P(r) [kDa]
from I(0) in P(r) [kDa]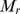 from sequence [kDa]
from sequence [kDa]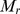 from I(0) in Guinier [kDa]
from I(0) in Guinier [kDa]