Figure 4.
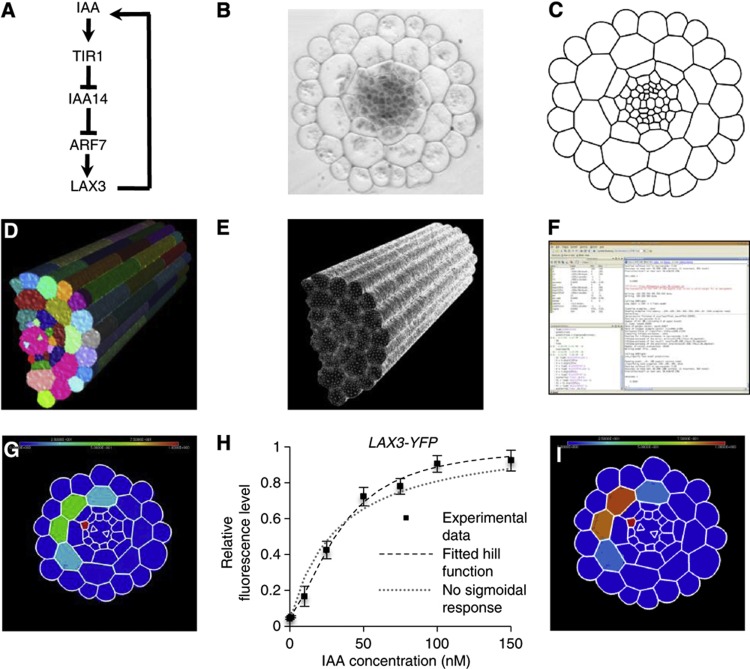
Computational 3D model of LAX3 induction by auxin in the cortical cells of the Arabidopsis root. (A) Representation of the gene regulatory network controlling lateral root emergence. IAA: auxin; TIR1: F-box protein that interacts with IAA14 to form the auxin receptor complex; IAA14: Aux/IAA protein acting as a negative regulator of ARF7; ARF7: Auxin response factor 7, a transcription factor that activates downstream auxin responsive genes; LAX3: auxin influx transporter. (B–F) The meshing pipeline. A cross-section of a resin embedded wild-type (Col-0) Arabidopsis root (B) was manually digitised using Adobe Illustrator (C) and then extruded to realistically recreate the third dimension (including transverse walls, see main text) (D). A triangular mesh of the 3D object surface was generated using the CGAL library (E) and the surface triangles were imported into MATLAB to run the simulations (F). (G) Model output showing diffuse LAX3 protein accumulation in four cortical cell files (as indicated by a heatmap scale ranging from 0 (blue) to 1 (red); simulations shown are for the case in which the auxin source is provided by cells in a single XPP file; indicated in red). (H) LAX3 auxin dose response was determined by confocal microscopy of the LAX3-YFP protein and fitted to a Hill function (best fit: Hill Coefficient=2; threshold=40 nM); if no sigmoidal response was assumed (in the sense that the Hill Coefficient=1), then the fitted curve consistently overestimated the LAX3 response at low levels of auxin and underestimated it at high levels of auxin. Values are plotted as mean (±s.e., n=3). (I) Including the sigmoidal response increased the sharpness of the predicted LAX3 expression pattern, but still shows expression in four cortical cells. Scale bar is 25 μm (B).