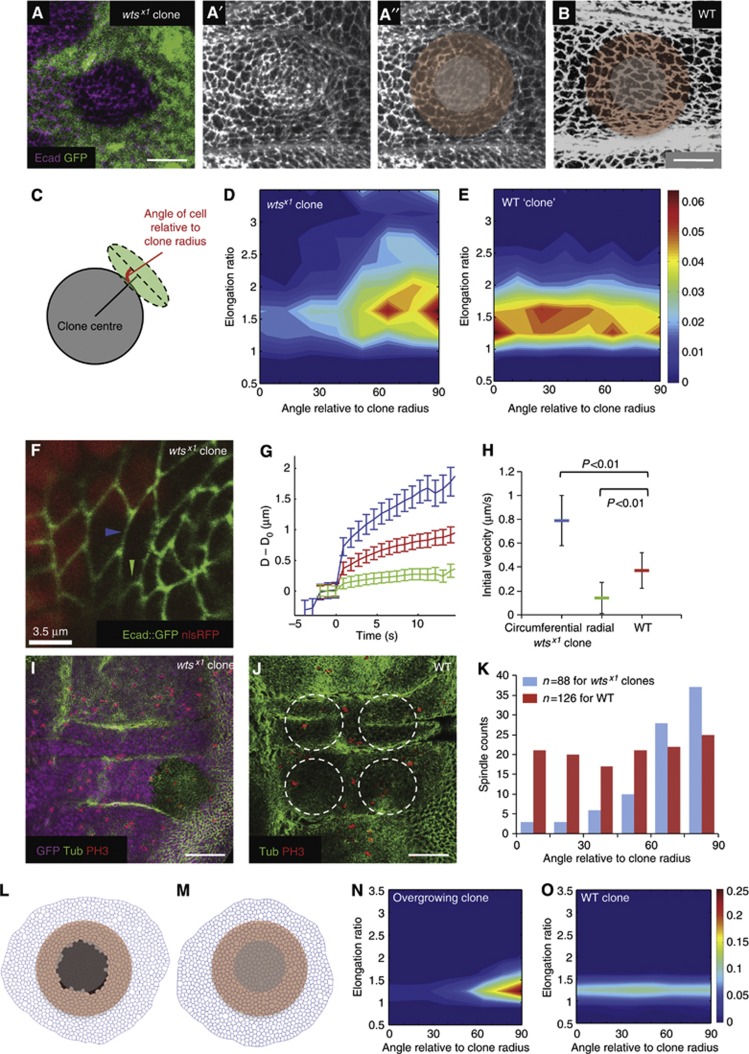
Figure 6.
The effect of altering proliferation rates in vivo. (A–A″) wts mutant clones marked by lack of nuclear GFP and stained for E-cadherin in the hinge region of the wing disc, scale=10 μm. A more basal GFP section is used to show the nuclear GFP signal. (A″) Schematic to represent the grey mutant cells removed from the cell-shape analysis and the surrounding red cells used in the analysis in (D). (B) The hinge region of a wild-type wing disc stained for E-cadherin, scale=10 μm. The red region is the corresponding control cells used for cell-shape analysis in (E). (C) Scheme of how the cell shapes around the clones are quantified (see Materials and methods). (D) Cell-shape analysis of cells surrounding wts clones (n=5 clones). Majority of cells around the clone are elongated circumferentially around the clone (tangential). (E) Cell-shape analysis of cells surrounding WT ‘clones’ (n=5 clones, each in the corresponding regions to each wts mutant clone). Cells are less elongated and show no specific orientation patterns. (F) A wts mutant clone marked by the absence of nuclear RFP and simultaneously expressing an E-cadherin::GFP transgene to allow live imaging of cell junctions for laser ablation. The blue arrowhead marks a typical circumferential junction of a wild-type cell bordering the mutant clone that was cut for the analysis (see Supplementary Movie 7). The green arrowhead marks a typical radial junction as used in the analysis. (G) Plot of increase in distance (μm) between the vertices of the cut junction (D−D0) against time (s) after laser cut, mean±s.e.m. Blue=circumferential junctions of wild-type cells surrounding wts mutant tissue, green=radial junctions of wild-type cells surrounding wts mutant tissue, red=wild-type hinge junctions. (H) The initial (maximum) recoil velocity of the vertices after the cut. Represented as mean±s.e.m. For circumferential junctions surrounding wts mutant tissue (blue), velocity=0.79±0.21 μm/s, n=39 junctions; for radial junctions surrounding wts mutant tissue (green), velocity=0.14±0.13 μm/s, n=30 junctions; for WT hinge junctions (red), velocity=0.37±0.15 μm/s, n=50 junctions. (I) wts mutant clone (lack of GFP) stained for tubulin and PH3 to identify mitotic spindle orientation, scale=50 μm. Only spindles close to clone boundaries are used for analysis in K (see Materials and methods). (J) A WT wing disc stained for tubulin and PH3, scale=50 μm. Circles show typical control ‘clone’ regions used for WT hinge spindle analysis. (K) Spindles surrounding wts mutant clones (blue) are oriented more circumferentially around the clone (tangential), whereas spindles in WT hinges show no orientation bias. (L) In silico simulation of an acute overgrowing clone (black cells) in wild-type tissue. (M) Control simulation where there is no acute overgrowing clone. (N) Cells surrounding the overgrowing clone (pink cells in L) elongate perpendicular to the clone radius. (O) Without acute overgrowth, cells show no elongation bias around the ‘clone’.