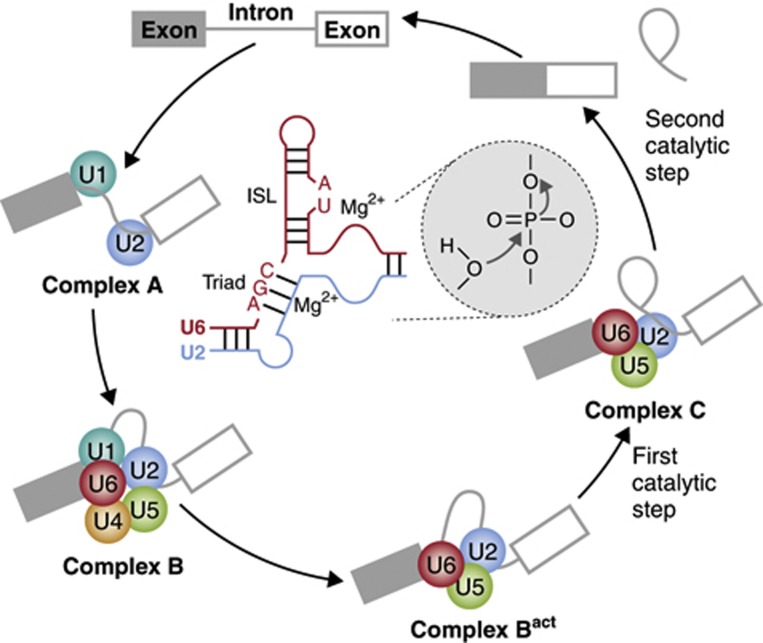
Figure 1.
Schematic representation of the process of Spliceosome assembly and of key RNA interactions at its core. Recognition of the 5’ splice site by U1 snRNP and/or the branch point region by U2 snRNP form complex A, which serves as a platform for assembly of the U4/5/6 tri-snRNP in complex B. Conformational rearrangements result in release of U1 and U4 and formation of the catalytically active Bact complex, in which the first step of the splicing reaction takes place, producing the free first exon and a lariat intron–exon intermediate. New conformational changes generate complex C, where exons 1 and 2 get ligated and the intron lariat released after the second catalytic step. Base pairing interactions between U2 and U6 snRNAs are critical for catalysis (central part). In particular, the AGC catalytic triad and an ISL in U6 help to coordinate magnesium ions that are believed to stabilize the nucleophile and leaving groups of the phosphates involved in each catalytic step.