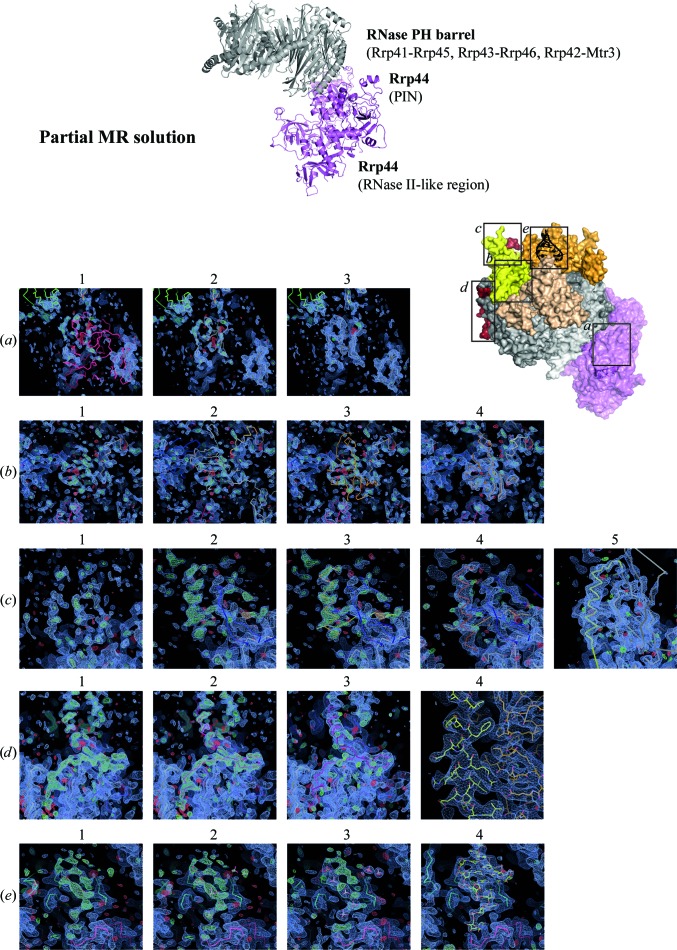
Figure 4.
Partial MR solution comprising about 60% of the complex (top). Manual placement of missing proteins was necessary, as it was not possible to obtain solutions for these domains by MR procedures. Interpretable positive density appeared as the model improved and became more complete from (a) to (e). (a) The CSD1 domain position of Rrp44 was offset by 12 Å, which could not be corrected by rigid-body refinement. Manual placement to the correct position and fine adjustment by rigid-body refinement resulted in much stronger electron density for this domain. (b) Positive density resembling a β-barrel was identified as belonging to the C-terminal domain of the Csl4 cap protein after superposing human Exo-9 on the partial structure. After a round of rigid-body refinement, electron density appeared with similar intensity as that of the neighbouring proteins. (c) The N-terminal domain of Csl4 was more difficult to discern, as the density was too large for the available model. Careful addition of backbone atoms revealed a more extended β-barrel fold than in the model. The α-helix turned out to belong to a region of the Rrp6 C-terminal tail. (d) Towards the end of model building, when all residues had been mutated to the yeast sequence and positional refinement had been employed, a curious density took shape on the surface of the Mtr3-Rrp43 subunits. After building a backbone and with a round of refinement, positive density for the side chain appeared. Using secondary-structure predictions of the unknown structure of the Rrp6 C-terminal tail together with good judgement of the chemical environment helped to place the side chains into the correct register. The final density for this region is shown and it indeed belonged to the C-terminal tail of Rrp6. (e) At the cap region, strong positive density suggested the possibility of an ordered ribonucleic acid chain. The phosphate ions placed into the strongest peaks turned out to be at distances typical of those of an RNA phosphate backbone. The electron density improved after a round of refinement, which allowed the placement of the respective ribose rings and bases. In fact, the initial positive density belonged to a strand of a duplex, as shown in the final model and the 2mF o − DF c map.