Summary
Memory T-cell repertoires are populated by clonotypes selected by an individual’s history of antigen exposures. Our previous analysis of middle-age CD8+ T-cell memory repertoires to the influenza-derived epitope M158-66, described a network of highly cross-reactive BV19 clonotypes responding to M158-66 and at least one peptide with a conservative amino acid substitution at either of two TCR contact positions. Here we report that some substitutions abrogate BV19 responses and favor responses with different BV. Cross-reactive T-cells using seven other BV families responded to 12 of 13 peptides tested. BV12 clonotypes define the most extensive cross-reactive network which encompasses seven peptides. We generated three-dimensional networks based on the peptides recognized and BV family used and observed a cluster of five peptides that includes M158-66 and another cluster of five peptides that does not include M158-66. The first cluster represents peptides structurally similar to M158-66, and the second represents peptides with more considerable changes in epitope recognition surface. We hypothesize that the second cluster represents the cross-reactive network around another unknown epitope or epitopes. This data supports a model of stable CD8+ T-cell memory networks that includes a substantial contribution from cross-reactive T-cells.
Keywords: Human, T-cells, T-cell receptor, Influenza, Cross-reactivity
Introduction
Immunological memory is a property of the adaptive immune system, which is composed of antigen-specific T and B cells that have participated in responses to a pathogen or vaccine. The population of cells that respond to a pathogen epitope defines a repertoire. Adult memory cell repertoires represent the history of antigen exposures that occurred in the individual and can be influenced by genetic and epigenetic factors as well as by innate response regulatory mechanisms [1].
An important question in human immunology asks how one memory T-cell repertoire is related to another. We hypothesize that a large part of the adult response can be made up of cross-reactive clonotypes. T-cell cross-reactivity is a recognized phenomenon in immunology [2-5] although its significance is still an open question. Because of the differences in thymic production and pathogen exposure history, a large portion of these cross-reactive clonotypes could be specific to each individual, which might make an analysis more difficult.
We have been investigating the adult T-cell repertoire in HLA-A2 individuals to the M158-66 epitope derived from the matrix protein of the influenza A virus, using the recall response to identify the repertoires. It has been shown that this recall response is dominated by T-cells bearing the BV19 (formerly BV17) gene segment [6] and a restricted IRSS amino acid motif in the CDR3 region of the TCR β chain[7-9]. Selection of M1-specific T-cells with these characteristics has been found to occur early in life [10]. Stimulation of PBMC from healthy adult blood donors with M158-66 and subsequent sequence analysis of BV19 TCR from CTL lines has revealed that the repertoires are polyclonal and complex [11, 12].
We have shown that M1 peptides with single amino acid substitutions at TCR contact residues can still be recognized by M158-66-specific BV19 clonotypes in vitro [13]. We found that the number of additional peptides recognized, i.e. the extent of crossreactivity, was as high as six and that the fraction of BV19 cross-reactive M158-66 clonotypes varied from 45% to 58% [13]. These cross-reactive clonotypes generated a fully connected network.
However, the response to M158-66 is not restricted to the BV19 family. Using BV19-depleted CTL, Lawson et al. [14] observed expansion of clonotypes using BV3, BV5s1, BV6, BV12, BV13, BV27, and BV28 in responses to M158-66. In this study we analyzed non-BV19 clonotypes that responded to either M158-66 and substituted peptides, or only to substituted M1 peptides. We found that 14 non-BV19 families can be involved in responses to M158-66 and/or to substituted peptides. Seven BV families were identified in response to M158-66, with four of them being cross-reactive. Of the other seven BV families, three were cross-reactive. We analyze the pattern of cross-reactivity in these data and discuss the results in terms of overlapping repertoires generated against different antigens.
Results
We have already described our choice of substitutions [13], which was based in part on the work of Gotch et al., [15] who used multiple substitutions at different positions of the M158-66 peptide to evaluate CTL function in vitro. The peptides sequences used are shown in Supporting Information Table 1. We have shown that some substitutions at TCR contact positions resulted in expansion of BV19 clonotypes that react with M158-66 and one or more of the substituted peptides. Such clonotypes defined the cross-reactivity of the M158-66 repertoire [13]. In this study we use the same three HLA-A2 individuals and focus on the cross-reactive clonotypes that do not use BV19 in response to the same panel of M1 peptides substituted at TCR contact residues.
To define specific responses we analyzed the CDR3 lengths of TCR BV cDNA amplicons from CD8 T-cells isolated from triplicate cultures stimulated with either M158-66 or one of the substituted peptides. Presence of a focused CDR3 length in at least two out of three cultures and its absence in no-peptide control cultures was considered specific (as an example data for BV12 are shown in Supporting Information Fig. 1).
Responses to M158-66 and substituted peptides not restricted to the BV19 family
For all three donors, 316 clonotypes were identified that responded to at least one of the 13 peptides and did not use BV19. These clonotypes used 14 other BV families to respond to one or more of the stimulating peptides. Out of 316 clonotypes, 20 were cross-reactive and used seven BV families in response to 12 of the peptides. One substituted peptide, A63, did not generate any cross-reactive clonotypes. The data relating to the cross-reactive clonotypes are summarized in Figure 1. The first three columns show the clonotype name, the BV family used, and the donor origin. For each BV family a different color is used and the peptide reactivity patterns are shaded to indicate data from different donors. For example, the M158-66 peptide was recognized by eight cross-reactive clonotypes. Five of the eight used BV18 (grey color) and the remaining three were each from a different family. We organized the peptides to optimize clustering of the data. The clustering will be important in later discussion. We summarize the responses by BV family in the two right columns.
Figure 1. Cross-reactive clonotypes responding to M158-66 and substituted peptides.
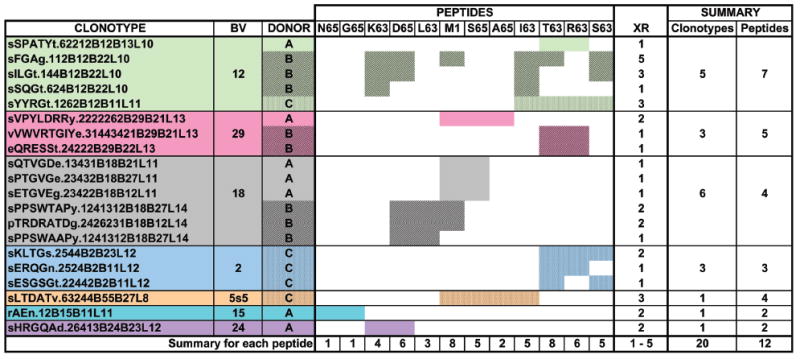
PBMC were cultured in triplicate with each peptide. TCR BV cDNA amplicons were fractionated by CDR3 length and a focused CDR3 length(s) in at least two out of three cultures used as an indicator of a specific response. Pooled data from the triplicate cultures for each peptide for all three donors are shown. Cross-reactive clonotypes that responded to the study peptides and do not use BV19 are identified in the left-most column. BV usage of the clonotypes is indicated in the next column, and each BV family is a separate color (green for BV12, pink for BV29, grey for BV18, pale blue for BV2, light brown for BV5s5, sky blue for BV15, and purple for BV24). Donors (third column from left) are indicated by different patterns and the same color/pattern is used to indicate response to the different peptides. The summary for each peptide is shown at the bottom. The column identified as XR shows the extent of cross-reactivity, which is calculated as the number of peptide-specific repertoires in which the clonotype was identified minus one. The XR range is indicated at the bottom. The number of clonotypes and peptides recognized for each BV family is in the two right-most columns, and is summarized at the bottom.
The extent of cross-reactivity (Fig. 1, column XR) reflects the number of peptides recognized by the same clonotype. By definition a cross-reactive clonotype has to recognize at least two peptides and the extent of cross-reactivity is one less than the number of peptides recognized. The maximum extent of cross-reactivity was five and the clonotype that showed this extent of cross-reactivity was found in Donor B repertoire, used BV12, and recognized M158-66. The highest extent of cross-reactivity for Donor C was three, shown by two clonotypes. The highest extent of cross-reactivity for Donor A was two.
Connectivity of non-BV19 repertoires
We have already described the use of graph theory for mapping out a cross-reactive BV19 network [13]. Peptides form the nodes of such a graph, and the cross-reactive clonotypes connect the various peptides. We refer to the connection as lines although they can also be referred to as edges.
BV12 network
BV12 forms the most extensive network of the cross-reactive clonotypes analyzed in this study (Fig. 2). There are seven peptides in the BV12 network, including M158-66. In the figure, the numbers in small circles indicate the number of cross-reactive clonotypes in the connection. The highest number of cross-reactive clonotypes in a connection is three (K63-I63). In a fully connected network, each peptide would make six connections, one to each of the other six peptides. However, this network is not fully connected due to R63 making connections with only three other peptides: I63, S63, and T63.
Figure 2. Connectivity of the BV12 repertoire specific for M158-66 and substituted peptides.
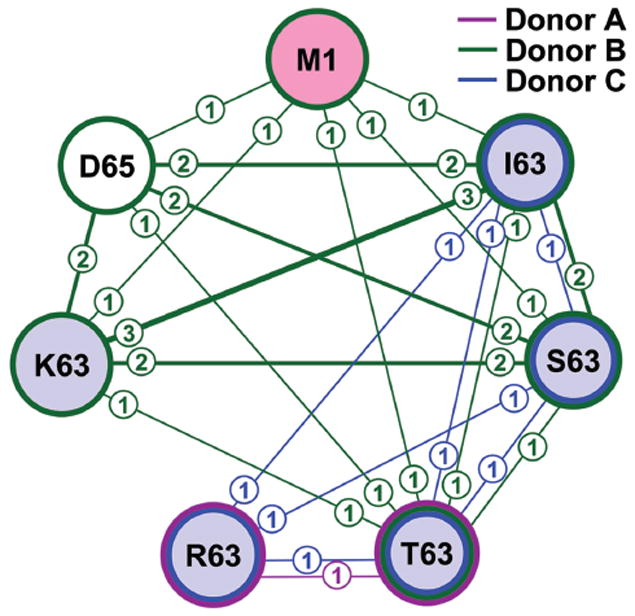
Large circles (nodes) identify the indicated peptides. The lines indicate a clonotype connection between any two peptides. The numbers in the circle associated with a line indicate the number of cross-reactive clonotypes in that connection; the thickness of each line also reflects this number. Colors inside circles indicate the position of the substitutions (pink for the original M158-66 peptide; pale blue for substitution at position 63; no color for substitution at position 65). Colors of the lines around each circle indicate the different donors (figure key). Pooled data from triplicate cultures for peptides that were recognized by BV12 clonotypes for all three donors are shown.
The contribution of each donor to the network is shown by using different colors for each line and around each peptide. It is evident that Donor B (green) contributes the most connections in this network, only missing connections to R63. The clonotype with the highest extent of cross-reactivity connects six peptides. Another clonotype connects four peptides: D65, I63, S63, and K63. A third clonotype connects only K63 and I63. Donor C has only one clonotype that connects four peptides, which do not include M158-66. Donor A has one clonotype that connects R63 to T63.
Other BV networks
Of interest are the six clonotypes in the BV18 network, which generate two donor-specific peptide recognition patterns. Three clonotypes from donor A recognize M1 and S65, whereas three from Donor B recognize L63 and D65, with two of these also recognizing M1 (Fig. 1). Each of the clonotype clusters shows some similarities in their CDR3 sequences; TGV in two Donor A clonotypes and PPSW in two donor B clonotypes. The other BV families form smaller networks.
Peptide-BV networks
To analyze the relationship between different BV and peptide networks, we performed cluster analysis by donor. To do so we present the peptide in each BV-defined cross-reactive network and connect the peptides (Fig. 3). For completeness we include our previous BV19 data [13]. Peptide footprints are in the same position on all BV layers and colored according to position of substitution. The top layer maps out the position of all the peptides. When the same peptide was present in different BV layers, we connected the same peptide footprint through the BV layers to create a three- dimensional network. For Donor A, the most connected peptides are M158-66 and S65, which were present on three BV layers: BV18, BV29, and BV19. A65 joined these peptides on the BV29 and BV19 layers. These three peptides were highly cross-reactive in the BV19 network described earlier [13]. On the other hand, D65 and K63 peptides on the BV24 layer were not connected with any other BV. From the BV12 layer (T63 and R63) only T63 was connected to another layer. For the BV15 layer, (G65 and N65), only G65 was connected to another layer. In both these cases the connection is to BV19. Two peptides on the BV19 layer (I63 and L63) were also not connected.
Figure 3. Cluster analysis of distribution of the cross-reactive clonotypes from different BV families identified in M158-66 and substituted peptide repertoires.
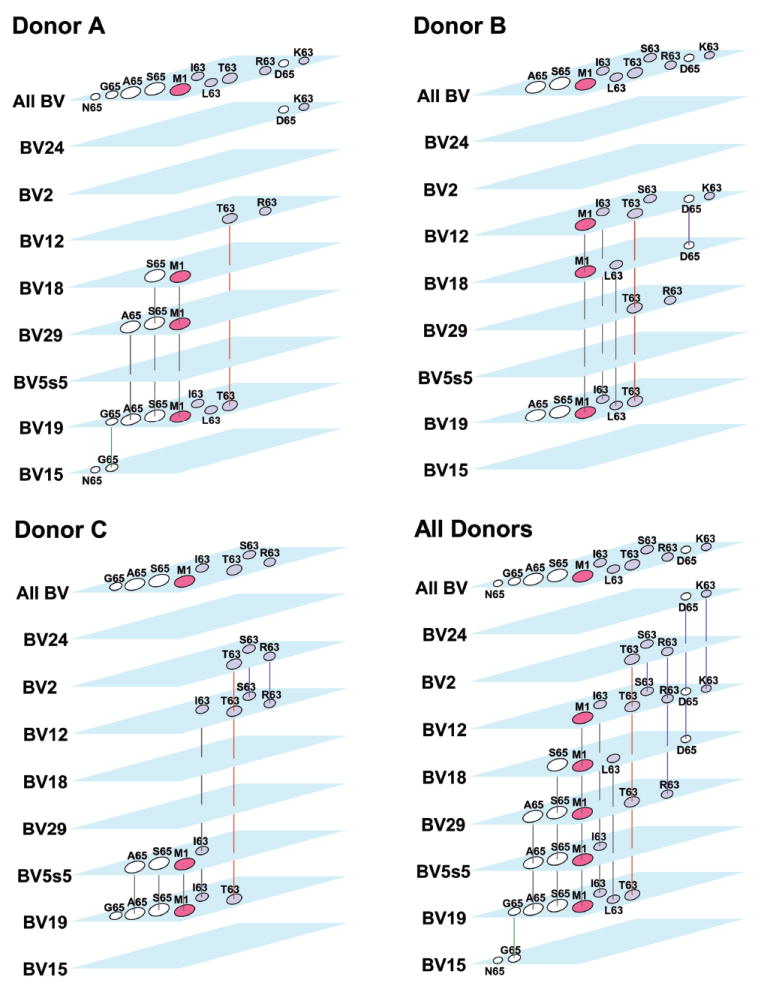
Each BV is shown as separate layer, with peptide footprints located at the same position. The total peptide footprint pattern is shown as “All BV” for ease of alignment. We connected peptides on different BV layers to make a three-dimensional network. The M1-S65-A65-I63-L63 cluster is connected with black lines. These peptides participated in the BV19 M1 network. T63-S63-R63 and D65-K63 clusters are connected by purple lines. T63 is connected with red lines to indicate its participation in two clusters. The G65-N65 cluster on BV15 connects to BV19 with a green line and represents an example of unusual cross-reactivity, as N65 was not cross-reactive with any other peptide. For each donor, Pooled data from triplicate cultures for each peptide and BV are shown. The bottom right panel combines data from all three donors.
The peptide-BV 3D network is donor specific. For example, for Donor B we did not observe the M1-S65-A65 peptide cluster on BV29 and BV18 as observed for Donor A. For Donor C however, the M1-S65-A65 cluster is recognized by BV5s5 utilizing clonotypes. L63 and I63 that were not connected in Donor A were connected to BV18 and BV12 in Donor B, and M158-66 showed the same pattern. T63 was as highly connected as M158-66 (three BV layers) and was cross-reactive in the BV19 network. From D65 and K63 on BV12, only D65 was connected with BV18. In Donor C I63 joined the M1 cluster on BV5s5 and was also observed on BV12, which is similar to Donor B. In Donor C we can also observe the T63-S63-R63 cluster that connects BV2 and BV12.
We combined data for all three donors and observed that all but one peptide are now connected between different BV layers. For example, the D65-K63 cluster on BV24 (Donor A) is now connected to BV12 and BV18 (Donor B). The T63-R63 cluster on the BV12 layer in Donors A and on BV29 in Donor B are now connected to each other as well as to BV2 and BV12 in Donor C. S63 on the BV12 layer in Donor B is now connected to BV2 and BV12 in Donor C. A cluster of four peptides M1-S65-A65-I63 now connects five BV layers: BV19, BV5s5, BV29, BV18, and BV12. Thus, there are three clusters of peptides that connect across BV and donors: M1-S65-A65-I63, T63-S63-R63, and D65-K63.
The M1-S65-A65-I63 cluster defined above belongs to the BV19 M1 network [13]. This network also includes L63, which was observed only in Donor B. To show the peptides that participate in the network first defined for BV19 and M1, all connections are shown in black. The connections for clusters T63-S63-R63 and D65-K63 also mentioned above are colored in purple. However, connections for T63 are colored in red to indicate its participation in the BV19 M1 network and the T63-S63-R63 cluster. The G65-N65 cluster on BV15 connects to BV19 through G65. This connection is colored in green, indicating its participation in the BV19 M1 network and the G65-N65 cluster.
Epitope-specific cross-reactive clonotype networks
The data presented above have mapped cross-reactive clonotypes and the peptides they respond to on the basis of BV usage. However, all the clonotypes present in the memory recall repertoire in an adult have been selected on pathogen exposure and the ensuing epitope-specific response. Therefore we divide our cross-reactive clonotype data on the basis of responding or not responding to M158-66, irrespective of BV usage.
The portion of the M1 cross-reactive network that uses BV19 has been described previously [13] and we extend it to include BV5s5, BV29, BV18, and BV12 clonotypes, which adds S63, K63, and D65 peptides (Fig. 4). This more extensive network is not fully connected as it was when it was restricted to BV19. Adding these new peptide nodes to the M1 network decreases connectivity while the number of cross-reactive clonotypes in the network is only increased slightly.
Figure 4. Connectivity of M158-66 repertoire for all BV families.
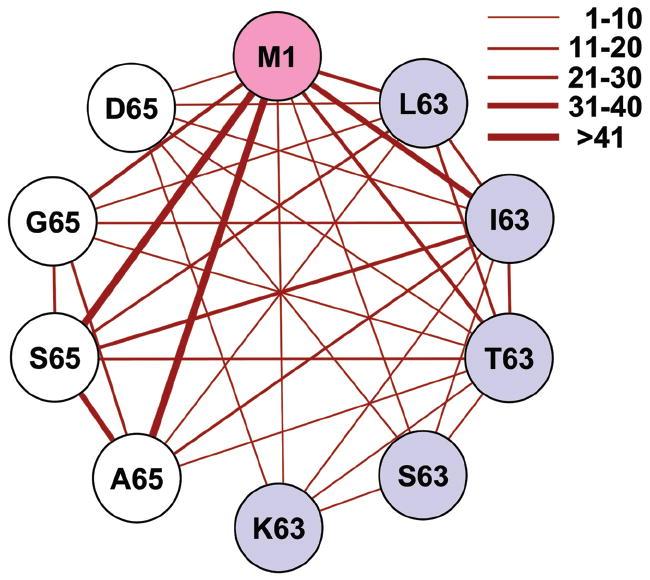
The thickness of the connecting lines reflects the number of clonotypes in each connection (range is shown in figure key). Number of clonotypes in each connection is omitted for simplicity. Colors inside circles indicate the position of the substitutions as described in legend for Fig. 2. Pooled data from triplicate cultures for each peptide and BV are shown. BV19 data from reference [13].
We propose that the cross-reactive clonotypes recognizing peptide clusters not connected to the M1 network constitute separate network(s) built around an unknown peptide or peptides (Fig. 5). This concept is further developed in the discussion.
Figure 5. Overlapping specificities of non-BV19 T-cell repertoires.
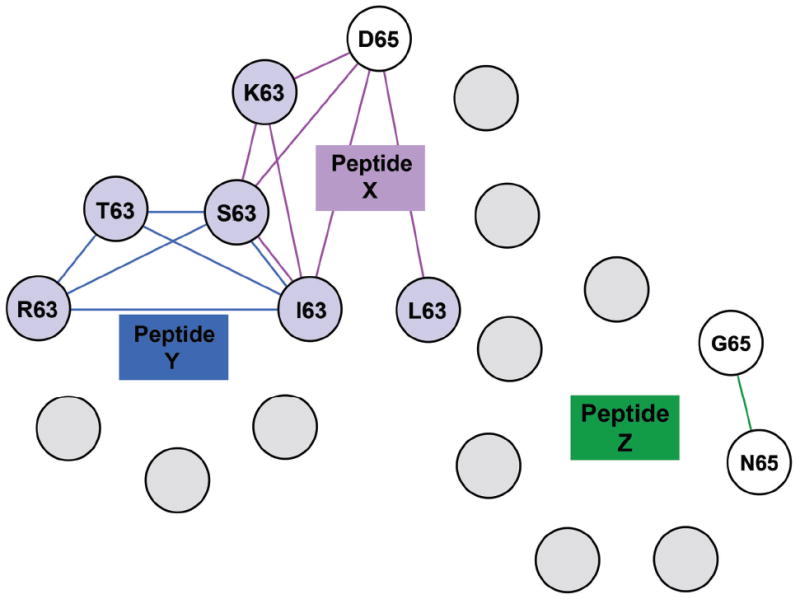
Each non-M158-66 cluster from Figure 3 was modeled as defining a separate cross-reactive network around a number of unknown epitopes. Colors inside circles indicate either the position of the substitutions as described in the legend for Figure 2 or unknown peptide (grey color). Colors of connecting lines correspond to the box color of Peptides X (purple), Y (blue), and Z (green). Some of these networks can be connected and others are unconnected to the others. All of these networks would also have connections to the M158-66 network but these are omitted for ease of viewing. Pooled data from triplicate cultures for each peptide and BV and all three donors are shown.
Discussion
We hypothesize that the adult human T-cell memory repertoire is composed of a network of clonotypes with overlapping specificities for pathogen-derived epitopes that have been encountered during the active phase of thymic output during infancy and childhood. This network then generates the emergent property of being able to respond to epitopes from many novel pathogens encountered during adult life with minimal morbidity or mortality. Starting with any one epitope and depending on the connectivity of the network around this epitope, as we change the structure of the peptide-MHC complex we would expect to lose the response of some clonotypes and gain the response of others. To test this expectation, we investigated the adult T-cell repertoire using the recall response to HLA-A2 restricted influenza A-derived M158-66 peptide as a model system. We have shown that BV19 responses to M158-66- and M1-substituted peptides are polyclonal with a high fraction of cross-reactive clonotypes that form a fully connected network [13]. Peptides participating in this network are structurally similar, hence the use of the same BV in their recognition.
In this study we analyzed non-BV19 cross-reactive responses in the same three donors. We identified cross-reactive clonotypes that used seven BV families to recognize 12 different peptides. Most of the peptides recognized by these non-BV19 clonotypes had more dramatic side-chain substitutions than those recognized in our previous analysis. The maximum extent of cross-reactivity among these clonotypes was five, one less than that observed for the BV19 network. While the extent of cross-reactivity was similar to the BV19 data, the fraction of cross-reactive clonotypes (20/316 = 6.3%) was much lower than the BV19 M158-66 clonotypes (97/190 = 51%). This decrease is due in part to the more extensive structural changes associated with the side-chain substitutions, changes that decreases the probability of recognition.
Peptides that generate cross-reactive clonotypes across different BV and different donors can be clustered (Fig. 3). The first cluster, M1-S65-A65-I63, is part of the M1 network (Fig. 4). A second cluster, R63-T63-S63, associated with BV2, BV12 and BV29 clonotypes, can define a different network, not containing M158-66. We propose that this network surrounds another epitope, Peptide Y (Fig. 5). The same can be done for a third cluster, K63-D65, associated with BV24, BV12 and BV18 clonotypes, and surrounding Peptide X. On the basis of clonotype cross-reactivity, both the X and Y networks would include I63 and S63. A third unconnected network around Peptide Z could contain clonotypes recognizing G65 and N65. We propose that the adult T-cell repertoire is populated by many networks similar to those described in this study. These networks would provide the ability to respond to novel epitopes.
Detection of T-cells that proliferate in response to substituted peptides raises the question of possible genetic variations of the M158-66 epitope, and thus the observed responses are actually naturally primed. RNA viruses like Influenza A, demonstrate a high mutation rate, which results in mutant viruses, or variants called quasispecies [16]. Kuroda et al., [17] used pyrosequencing to characterize the quasispecies of pandemic 2009 influenza A virus (A/H1N1/2009). They detected several mutants for M158-66 epitope, one of which resulted in substitution of Val at position 63 for Gly. Thus, we cannot exclude generation of mutant M1 viruses upon infection, which might results in presentation one of the peptides we studied here. Because most of our substitutions are nonconservative we consider this unlikely. However, this is an interesting question.
It is important to know how our in vitro system reflects events that occur in vivo. Infection causes a complex immune response that includes processing of multiple epitopes and feedback based on the extent of pathogen inflicted damage. The feedback takes the form of factors that include cell surface molecules and cytokines. These factors can set the threshold of the T-cell responses, perhaps tuning cross-reactivity. Our in vitro system minimizes this complexity because we add only one peptide and only supplement with IL-2. While IL-2 might change the threshold for TCR signaling and affect the extent of cross-reactivity, our data show that under these relatively simple in vitro conditions cross-reactivity can be measured.
Our analysis of position 63 and 65 was incomplete since we only covered 14 of 40 possible side chains at these two positions. We chose our substituted peptides based in part on the work of Gotch et al. [15], who evaluated different M158-66 substitutions on CTL function in vitro and showed that this general approach could work. This restriction in the peptide usage set limits on generalization from our data. The average cross-reactivity is 6% for non-BV19 clonotypes for the all three donors and all the peptides tested. If we generalize these results under the assumption that the untested peptides would bind to HLA-A2, generate some level of response, and none would be cross-reactive, our value for the cross-reactivity for this particular sequence space would be ~2% (14/40 * 6%). This would represent the lower limit of the fraction of cross-reactivity that might be expected in the repertoire. However, if one of the untested substitutions would bring the pHLA-A2 structure closer to one of the unknown peptides, the percentage might increase.
There are a few examples of networks described in the literature, predominantly in Ab responses. Cross-reactive plasma-cell-derived anti-HA antibodies from either H3N2 infected or TIV vaccinated subjects were able to bind up to six HA subtypes [18]. Anti-HA antibodies from H3N2-infected subjects neutralized both H1N1 and H3N2 viruses. A network of cross-reactivity among MSP3-family members of Plasmodium falciparum malaria provides another example [19]. In this example, human antibodies affinity-purified against each of the C-terminal recombinant proteins were found to display varying degrees of cross-reactivity with the remaining members of the MSP3 family.
Our network model extends the idea of cross-reactive heterologous immunity [5, 20]. The approach of stimulation with substituted peptides could be extended to generate cross-reactive epitope maps of complex memory repertoires such as found in man. To the extent that other previously encountered epitopes generate similar cross-reactive repertoires, we would expect that these repertoires will also be able to respond to new pathogens. Such cross-reactive clonotypes could also provide for coverage of escape variants.
Materials and methods
Peptides
Influenza A matrix M1 58-66 peptide and all substituted peptides (sequences are shown in Supporting Information Table 1) were synthesized by standard solid-phase methods, purified by HPLC, and confirmed by mass spectrometry (Peptide Core, Blood Research Institute, BloodCenter of Wisconsin).
CTL isolation, RNA isolation, cDNA preparation
PBMC were collected from buffy coats of three healthy HLA-A2.1 donors (age 37, 38, and 47) using lymphocyte separation medium (Cellgro, Mediatech, Manassas, VA). Prior to use, they were cryopreserved in human AB serum containing 10% DMSO at a concentration of 107/ml with a control freezing system (CryoMed Freezer, Fisher Scientific, Pittsburg, PA). All samples were obtained under Institutional Review Board-approved protocols (IRB# BC 05-11 and BC 04-22). Frozen PBMC were stored in liquid nitrogen at -180°C. To establish CTL, PBMC were thawed and placed into complete RPMI supplemented with 10% heat-inactivated human AB serum (Atlanta Biologicals, Lawrenceville, GA) at room temperature. PBMC were cultured at 2 × 106 cells/2 ml in the presence of 1 μM of influenza A matrix peptide M158-66 or one of the substituted peptides and 10 U/ml of recombinant IL-2 in 12-well plates for 7 days. Additional recombinant IL-2 (10U/ml) was added on day 4. All cultures were established in triplicate. No peptide culture was used as negative control. Primed cells were restimulated in the presence of recombinant IL-2 (10 U/ml) and an equal number of irradiated (3000 R) autologous PBMC that had been preloaded with M158-66 or a corresponding substituted peptide. After 2 weeks in culture, CD8+ T-cells were separated using Dynal® CD8 Positive Isolation Kit (Invitrogen, Carlsbad, CA). RNA isolation was performed using Dynabeads® mRNA DIRECT™ Kit (Invitrogen). For Donor C, RNA from cultures stimulated with peptides substituted at position 63 was isolated after 3 weeks of culturing with no CD8 isolation. For all donors, cDNA was synthesized immediately after RNA isolation using oligo(dT) (5’ TTTTTTTTTTTTTTTTTT 3’) primer and Moloney murine leukemia virus reverse transcriptase (Invitrogen).
CDR3 spectratyping
BV genes were amplified from cDNA using 24 BV family-specific primers and fluorochrome-labeled TCR CB primer as described [21]. 1 μl of each PCR product was used for fragment analysis (3100 Genetic Analyzer, Applied Biosystems, Foster City, CA). Data analysis was performed using proprietary software (Flynn Creek Biosciences, Hubertus, WI).
PCR product cloning, sequencing, and colony counting
10 μl of PCR product was used for electrophoresis on a 5% polyacrylamide gel. Specific bands were visualized by fluorescence detection (Typhoon TRIO+, GE Healthcare Life Sciences, Piscataway, NJ) and excised; alternatively, the PCR product was directly subcloned. DNA extraction was performed using QIAEX® II Gel Extraction Kit (Qiagen, Valencia, CA). 10 μl of extracted DNA was amplified for three cycles and subcloned using TOPO TA Cloning® Kit for Sequencing (Invitrogen). Competent cells were inoculated on Luria-Bertani agar plates (Difco™ LB Agar, Miller, BD, Franklin Lakes, NJ) containing ampicillin (100 μg/ml) and incubated overnight at 37°C. Twenty-four colonies for each sample were randomly chosen for inoculation in 400 μl Luria-Bertani broth (Difco LB Broth, Miller, BD) containing ampicillin (100 μg/ml) in deep 96-well plates and incubated overnight on a shaker (250 rpm) at 37°C. 160 μl of bacterial broth was frozen at -80°C in the presence of 40 μl of 50% glycerol. Sequences were performed by AGENCourt Bioscience Corporation (Beckman Coulter Company, Beverly, MA) or GeneWiz, Inc. (South Plainfield, NJ). Data were analyzed using FinchTV software (Geospiza, Inc., Seattle, WA). Clonotypes were named on the basis of their amino acid sequence with a numerical coding that allows reconstruction of the nucleotide sequence [22].
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health (Grant U19 AI062627).
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Ahmed R, Rouse BT, editors. Immunological memory. Immunol Rev. 2006;211:5–337. [Google Scholar]
- 2.Wucherpfennig KW. T cell receptor crossreactivity as a general property of T cell recognition. Mol Immunol. 2004;40:1009–1017. doi: 10.1016/j.molimm.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Holler PD, Kranz DM. T cell receptors: affinities, cross-reactivities, and a conformer model. Mol Immunol. 2004;40:1027–1031. doi: 10.1016/j.molimm.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 4.Wilson DB, Wilson DH, Schroder K, Pinilla C, Blondelle S, Houghten RA, Garcia KC. Specificity and degeneracy of T cells. Mol Immunol. 2004;40:1047–1055. doi: 10.1016/j.molimm.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 5.Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses. Immunol Rev. 2010;235:244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehner PJ, Wang EC, Moss PA, Williams S, Platt K, Friedman SM, Bell JI, Borysiewicz LK. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. J Exp Med. 1995;181:79–91. doi: 10.1084/jem.181.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss PA, Moots RJ, Rosenberg WM, Rowland-Jones SJ, Bodmer HC, McMichael AJ, Bell JI. Extensive conservation of alpha and beta chains of the human T-cell antigen receptor recognizing HLA-A2 and influenza A matrix peptide. Proc Natl Acad Sci U S A. 1991;88:8987–8990. doi: 10.1073/pnas.88.20.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naumov YN, Hogan KT, Naumova EN, Pagel JT, Gorski J. A class I MHC-restricted recall response to a viral peptide is highly polyclonal despite stringent CDR3 selection: implications for establishing memory T cell repertoires in “real-world” conditions. J Immunol. 1998;160:2842–2852. [PubMed] [Google Scholar]
- 9.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 10.Lawson TM, Man S, Williams S, Boon AC, Zambon M, Borysiewicz LK. Influenza A antigen exposure selects dominant Vbeta17+ TCR in human CD8+ cytotoxic T cell responses. Int Immunol. 2001;13:1373–1381. doi: 10.1093/intimm/13.11.1373. [DOI] [PubMed] [Google Scholar]
- 11.Naumov YN, Naumova EN, Hogan KT, Selin LK, Gorski J. A fractal clonotype distribution in the CD8+ memory T cell repertoire could optimize potential for immune responses. J Immunol. 2003;170:3994–4001. doi: 10.4049/jimmunol.170.8.3994. [DOI] [PubMed] [Google Scholar]
- 12.Naumov YN, Naumova EN, Clute SC, Watkin LB, Kota K, Gorski J, Selin LK. Complex T cell memory repertoires participate in recall responses at extremes of antigenic load. J Immunol. 2006;177:2006–2014. doi: 10.4049/jimmunol.177.3.2006. [DOI] [PubMed] [Google Scholar]
- 13.Petrova GV, Naumova EN, Gorski J. The polyclonal CD8 T cell response to influenza M158-66 generates a fully connected network of cross-reactive clonotypes to structurally related peptides: a paradigm for memory repertoire coverage of novel epitopes or escape mutants. J Immunol. 2011;186:6390–6397. doi: 10.4049/jimmunol.1004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson TM, Man S, Wang EC, Williams S, Amos N, Gillespie GM, Moss PA, Borysiewicz LK. Functional differences between influenza A-specific cytotoxic T lymphocyte clones expressing dominant and subdominant TCR. Int Immunol. 2001;13:1383–1390. doi: 10.1093/intimm/13.11.1383. [DOI] [PubMed] [Google Scholar]
- 15.Gotch F, McMichael A, Rothbard J. Recognition of influenza A matrix protein by HLA-A2-restricted cytotoxic T lymphocytes. Use of analogues to orientate the matrix peptide in the HLA-A2 binding site. J Exp Med. 1988;168:2045–2057. doi: 10.1084/jem.168.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domingo E, Baranowski E, Ruiz-Jarabo CM, Martin-Hernandez AM, Saiz JC, Escarmis C. Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis. 1998;4:521–527. doi: 10.3201/eid0404.980402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda M, Katano H, Nakajima N, Tobiume M, Ainai A, Sekizuka T, Hasegawa H, et al. Characterization of quasispecies of pandemic 2009 influenza A virus (A/H1N1/2009) by de novo sequencing using a next-generation DNA sequencer. PLoS One. 2010;5:e10256. doi: 10.1371/journal.pone.0010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, et al. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One. 2011;6:e25797. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh S, Soe S, Weisman S, Barnwell JW, Perignon JL, Druilhe P. A conserved multi-gene family induces cross-reactive antibodies effective in defense against Plasmodium falciparum. PLoS One. 2009;4:e5410. doi: 10.1371/journal.pone.0005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornberg M, Clute SC, Watkin LB, Saccoccio FM, Kim SK, Naumov YN, Brehm MA, et al. CD8 T cell cross-reactivity networks mediate heterologous immunity in human EBV and murine vaccinia virus infections. J Immunol. 2010;184:2825–2838. doi: 10.4049/jimmunol.0902168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yassai M, Naumova EN, Gorski J. Generation of TCR spectratypes by multiplex PCR for T cell repertoire analysis. In: Oksenberg J, editor. The human antigen T cell receptor: selected protocols & applications. Landes Company and Chapman & Hall; Austin, TX: 1997. pp. 327–372. [Google Scholar]
- 22.Yassai MB, Naumov YN, Naumova EN, Gorski J. A clonotype nomenclature for T cell receptors. Immunogenetics. 2009;61:493–502. doi: 10.1007/s00251-009-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


