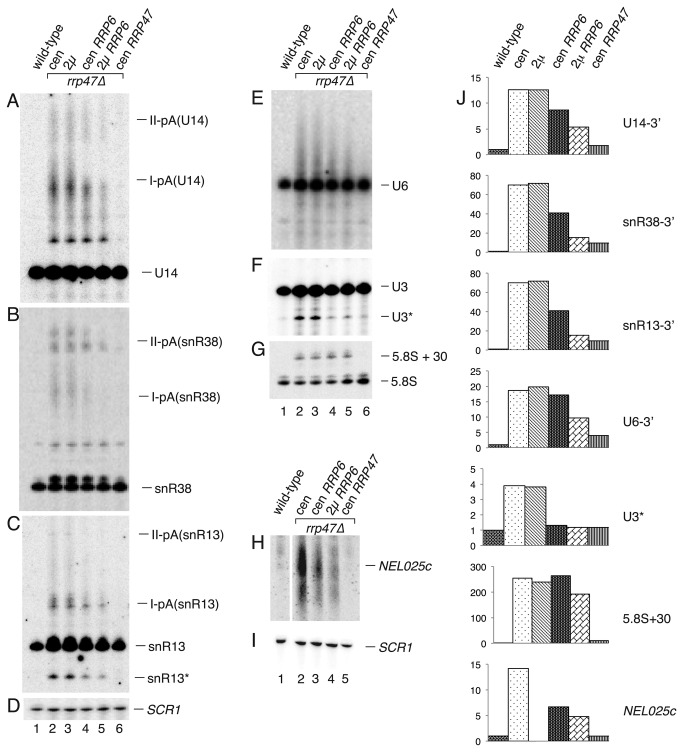
Figure 5. Rrp6 overexpression suppresses RNA phenotypes in rrp47∆ mutants.
Northern analyses of total cellular RNA isolated from a wild-type strain, an isogenic rrp47∆ mutant and from rrp47∆ mutants expressing exogenous Rrp6 or Rrp47 from either centromeric (cen) plasmids or 2 micron-based (2μ) constructs. RNA was resolved through 8% denaturing acrylamide gels, transferred to nylon membranes and hybridised with probes complementary to specific RNAs, as follows: (A) U14; (B) snR38; (C) snR13; (D) SCR1; (E) U6; (F) U3; (G) 5.8S; (H) NEL025c; (I) SCR1. Blots shown in A-G and H-I are from distinct gels. Dispersed bands labelled I-pA and II-pA in panels A-C represent snoRNAs that are polyadenylated after termination at sites I and II, respectively. The bands labelled snR13* and U3* are 5’ truncated forms of snR13 and U3. (J) Quantification of signals for the 3’ extended forms of U14, snR38, snR13, U6 and 5.8S, the truncated U3 RNA and the NEL025c mRNA are shown for each strain. Average values of two data sets are normalised to SCR1 loading controls and expressed relative to the level of the RNA observed in the wild-type strain.