A novel tissue culture model for studying cellular constriction is introduced and used to show that activation of the Fog signaling pathway depends on Ric-8 and that Ric-8 preferentially binds and localizes inactive Cta. Conserved residues are identified within Ric-8 that are important for the binding and function of Cta.
Abstract
Heterotrimeric G proteins, composed of α, β, and γ subunits, are activated by exchange of GDP for GTP on the Gα subunit. Canonically, Gα is stimulated by the guanine-nucleotide exchange factor (GEF) activity of ligand-bound G protein–coupled receptors. However, Gα subunits may also be activated in a noncanonical manner by members of the Ric-8 family, cytoplasmic proteins that also act as GEFs for Gα subunits. We used a signaling pathway active during Drosophila gastrulation as a model system to study Ric-8/Gα interactions. A component of this pathway, the Drosophila Gα12/13 subunit, Concertina (Cta), is necessary to trigger actomyosin contractility during gastrulation events. Ric-8 mutants exhibit similar gastrulation defects to Cta mutants. Here we use a novel tissue culture system to study a signaling pathway that controls cytoskeletal rearrangements necessary for cellular morphogenesis. We show that Ric-8 regulates this pathway through physical interaction with Cta and preferentially interacts with inactive Cta and directs its localization within the cell. We also use this system to conduct a structure–function analysis of Ric-8 and identify key residues required for both Cta interaction and cellular contractility.
INTRODUCTION
G protein–coupled receptors (GPCRs) are a highly conserved family of transmembrane receptors that evolved to detect a wide range of signals, including neurotransmitters, hormones, odorants, and light. These receptors have a characteristic topology that spans the membrane via seven α-helices and are oriented with their N-termini toward the extracellular space, their C-termini inside the cell, and three interhelical loops on each side. Ligand binding allows the cytoplasmic domains of the GPCR to activate heterotrimeric G-proteins, downstream signaling molecules that consist of a GTP-binding α subunit that exists in 1:1:1 stoichiometry with a β and a γ subunit. These three proteins form a tightly bound inactive heterotrimer when Gα is in its GDP-bound state. Activation of the GPCR induces a conformational change that triggers its guanine nucleotide exchange factor (GEF) activity for Gα, causing Gα to exchange bound GDP for GTP. Active Gα-GTP dissociates from the Gβγ heterodimer, and both species are able to regulate downstream effector molecules, such as ion channels and enzymes that produce second messengers. Gα subunits have an intrinsic GTPase activity that hydrolyzes GTP to GDP, causing the complex to reform into its inactive state. This cycle of activation and inactivation may be modulated by accessory factors, such as regulator of G protein signaling (RGS) proteins that accelerate the rate of GTP hydrolysis by Gα subunits (for review see Rossman et al., 2005; Siderovski and Willard, 2005; Oldham and Hamm, 2008). Thus, although the core regulatory component in heterotrimeric G protein signaling is the nucleotide-bound state of the Gα subunit, the activities of these molecules are affected by accessory factors that may reflect various signaling inputs into the pathways.
At the biochemical level Ric-8 acts as a noncanonical GEF for multiple families of Gα subunits (Tall et al., 2003; Chan et al., 2011) by associating with Gα-GDP, often complexed with a guanine-nucleotide dissociation inhibitor (GDI), such as the Go-Loco repeat–containing family of proteins (e.g., GPR1/2 in Caenorhabditis elegans and Pins in Drosophila melanogaster). Ric-8 binding inactive Gα facilitates GDP release and promotes the formation of a transient nucleotide-free state, which allows Gα-GTP exchange by cytosolic excess of GTP (Tall et al., 2003). In addition, Ric-8 binds to and drives dissociation of Gα-GDP complexed with a GDI, subsequently freeing Gα to engage other effectors (Hampoelz et al., 2005; Tall and Gilman, 2005; Thomas et al., 2008). Recently Ric-8 was identified as a chaperone involved in the biosynthesis of mammalian Gα subunits and their subsequent localization to the plasma membrane (Gabay et al., 2011). Thus several lines of evidence indicate that Ric-8 regulates multiple aspects of Gα function.
A growing body of data has implicated the Ric-8 family of proteins as important accessory molecules involved in heterotrimeric G protein signaling in a variety of developmental processes (for review see Hinrichs et al., 2012). Ric-8 is a highly conserved cytosolic protein that was originally identified in a screen for proteins required for Gαq signaling in the C. elegans nervous system (Miller et al., 2000). Since then, Ric-8 has been implicated as a regulator of signaling in events as diverse as fungal pathogenesis and development (Li et al., 2010; Wright et al., 2011) to modulation of mammalian vision, taste, olfaction, and bone formation (Tõnissoo et al., 2003, 2010; Von Dannecker et al., 2005, 2006; Dhingra et al., 2008; Kerr et al., 2008; Fenech et al., 2009; Yoshikawa and Touhara, 2009; Grandy et al., 2011; Maldonado-Agurto et al., 2011). Ric-8 plays a well-defined role in spindle orientation during mitosis of asymmetrically dividing cells. During early divisions of the C. elegans embryo Ric-8 acts through Gαi family members to establish the position of the mitotic spindle through modulation of pulling forces along the anterior–posterior axis (Miller and Rand, 2000; Afshar et al., 2004, 2005; Couwenbergs et al., 2004). Similarly, in Drosophila, Ric-8 functions through Gαi to align the mitotic spindle in both neuroblast and sensory organ precursor cells (David et al., 2005; Hampoelz et al., 2005; Wang et al., 2005). Recent findings also show that Ric-8 is important for spindle alignment in asymmetric cell division in mammalian tissue culture (Woodard et al., 2010). In addition to spindle positioning, Ric-8 regulates cytoskeletal rearrangements during dorsal ruffle formation via Gα13 in mammalian tissue culture (Wang et al., 2011). These data demonstrate that Ric-8, through its interaction with Gα subunits, functions to regulate diverse processes during G protein signaling events, including cytoskeletal behavior.
Drosophila gastrulation is a powerful model system with which to study heterotrimeric G protein signaling within a developmental context. During this process, the Drosophila blastoderm undergoes a series of highly orchestrated cell movements to drive subsets of cells into the interior of the embryo to establish the germ layers. One of the hallmarks of gastrulation is the invagination of a subset of epithelial cells along the ventral midline to form a structure called the ventral furrow (Leptin, 1995). Furrow formation is driven by concerted cellular shape changes in which apical constriction of the actin network by myosin II has the net effect of driving the internalization of the mesodermal precursor cells (Dawes-Hoang et al., 2005; Martin et al., 2009). Genetic analysis of this pathway has identified several components believed to act sequentially to trigger apical constriction. First, the midline epithelial cells destined to invaginate secrete an extracellular protein, Folded gastrulation (Fog), from their apical domains. Fog acts as an autocrine signal and binds to an unidentified transmembrane receptor that then signals through a heterotrimeric G protein complex containing the Drosophila Gα12/13 subunit, Concertina (Cta; Costa et al., 1994; Morize et al., 1998). Mutations in the Gβ13F and Gγ1 subunits exhibit gastrulation defects and, presumably, comprise the βγ subunits of the heterotrimer along with Cta (Wang et al., 2005). Cta activates a guanine nucleotide exchange factor, RhoGEF2, relocalizing RhoGEF2 from the plus-end tips of growing microtubules to the cortex, where it is docked by its interaction with a transmembrane protein, T48 (Rogers et al., 2004; Nikolaidou and Barrett, 2004; Kölsch et al., 2007). RhoGEF2 then activates the small G protein Rho1, which activates myosin II at the apical domain via Rho kinase (Rok), thus producing contraction (Barrett et al., 1997; Nikolaidou and Barrett, 2004; Dawes-Hoang et al., 2005). Mutations in any Fog pathway component interfere with the timing or execution of normal gastrulation. This pathway also has been implicated in epithelial remodeling during later stages of development (Nikolaidou and Barrett, 2004). Drosophila gastrulation events are highly analogous to epithelial remodeling in other multicellular organisms, most notably neural tube formation in the developing vertebrate embryo, and downstream signaling components are conserved between invertebrates and vertebrates (Lecuit and Lenne, 2007). Thus we are using the Drosophila Fog signaling pathway as a model system to investigate general mechanisms of signaling during tissue remodeling.
Given the central importance of Cta to Drosophila gastrulation, it is a useful model to study potential interactions between Ric-8 and Gα12/13-class subunits. Previous studies showed that Ric-8 mutants exhibit gastrulation defects that resemble Cta loss of function (Hampoelz et al., 2005; Wang et al., 2005). Further, Ric-8 was recently shown to regulate the cortical stability of actin in cells of the ventral furrow undergoing apical constriction (Kanesaki et al., 2013). However, the mechanistic details through which Ric-8 functions in this process remain to be determined. Here we examine the role of Ric-8 in the Fog pathway using a novel cell-based assay for Fog-induced cellular contractility. Using RNA interference (RNAi), we show that Ric-8 is necessary for Fog signaling and that it functions within the pathway at the level of Cta. We also find that Ric-8 directly interacts with Cta and exhibits higher affinity for inactive Cta mutants (GTP-free). We present biochemical data that show that Ric-8 preferentially binds and specifically acts to localize inactive Cta downstream of Fog/GPCR signaling. Finally, we conduct a mutagenesis screen of conserved charged amino acid residues and identify specific residues within Ric-8 required for Cta function and/or establishment of a binding interface between the two molecules. On the basis of our results, we propose a model in which Ric-8 acts downstream of Fog pathway activation to localize/scaffold inactive Cta, potentiating Fog signaling to drive persistent cellular constriction
RESULTS
Reconstitution of Fog-stimulated cellular contractility in a cultured cell model
To study the effect of Fog signaling on cell morphology, we developed a cell culture system to allow us to replicate in vivo signaling events. We began by engineering a stable S2 cell line that expresses full-length Fog tagged at its C-terminus with the Myc epitope under an inducible metallothionein promoter (S2:Fog-Myc). Costa et al. (1994) originally hypothesized that Fog is a secreted protein based on hydropathy analysis of the protein's primary sequence, which revealed the presence of an N-terminal 12–amino acid hydrophobic region predicted to function as a signal sequence. Later analysis of Fog localization in cells of the embryonic ventral furrow and posterior midgut showed that the protein localized to membrane-bound organelles targeted for the apical surface of the blastoderm epithelia (Dawes-Hoang et al., 2005). To test whether Fog is secreted from S2:Fog-Myc cells, we induced its expression with copper sulfate for 48 h, collected the conditioned medium, and concentrated it ∼20-fold. An affinity-purified antibody against the N-terminus of Fog recognized a single protein with a molecular weight of ∼150 kDa on immunoblots of conditioned medium from induced S2:Fog-Myc cells, and the same-sized band was also recognized by a monoclonal anti-Myc antibody. Neither antibody recognized the protein in conditioned medium collected from untransfected S2 cells (Figure 1A). Thus, as found in tissues in the Drosophila blastoderm preceding cellular shape change, ectopic Fog-Myc is expressed in S2 cells as a secreted protein.
FIGURE 1:
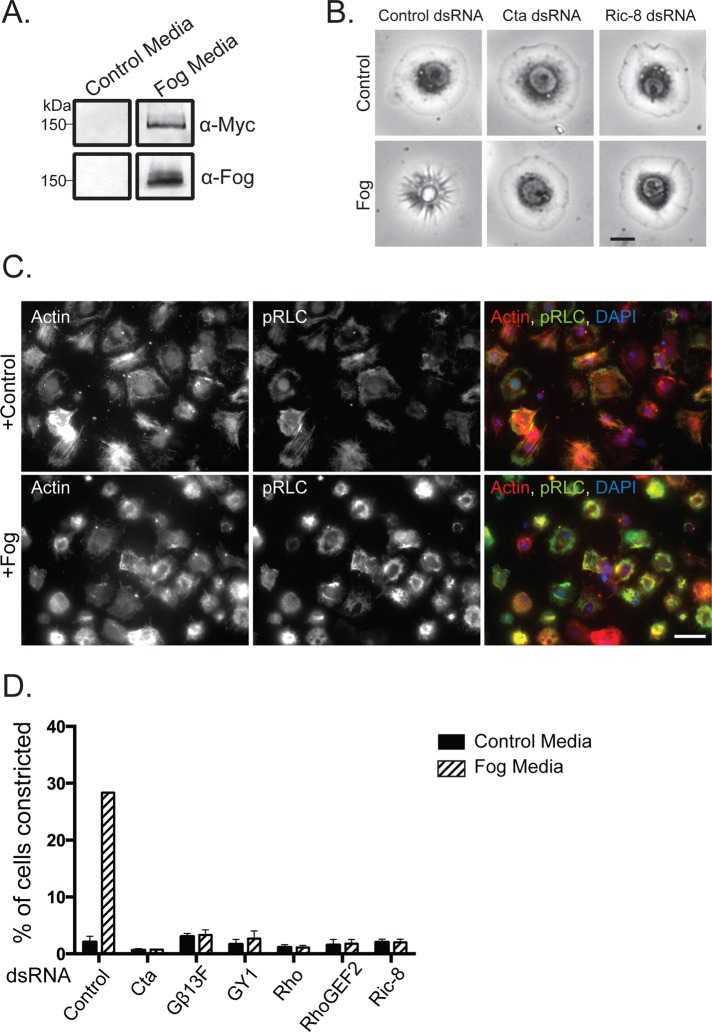
Recapitulation of Fog signaling in S2R+ cells. (A) Fog-Myc is secreted into the medium of a stable cell line expressing the construct but not by untransfected control S2 cells. Fog-Myc is recognized by anti-Myc and anti-Fog by immunoblot. (B) S2R+ cells undergo cellular shape changes in response to ectopic Fog application. RNAi-mediated depletion of Cta or Ric-8 prevents Fog-induced cellular constriction. Scale bar, 10 μm. (C) Fog-induced S2R+ contraction is accompanied by an increase in active phosphorylated nonmuscle myosin II (pRLC). S2R+ cells were treated with either control or Fog-containing media and stained for actin (red), pRLC (green), and DNA (blue). Scale bar, 100 μm. (D) S2R+ cells lose their responsiveness to Fog after RNAi against known pathway components, as well as Ric-8. Percentage of cells constricting in response to Fog was measured within a population of cells (±SEM).
We next screened an assortment of immortalized Drosophila cell lines for their ability to respond to Fog-conditioned medium. Previously we showed that activation of the Rho1 pathway in S2 cells caused the cells to adopt a contracted morphology (Rogers et al., 2004). We therefore used this read-out to test the ability of S2 cells, S2-receptor + (S2R+) cells, and several immortalized lines derived from imaginal discs to respond to Fog. S2R+ cells are a subline derived from S2 cells that express receptors not found in S2 cells (Yanagawa, 1998). Neither S2 cells nor the other epithelial lines we tested changed their shape in response to Fog perfusion (unpublished data). However, S2R+ cells exhibited a robust morphological response upon perfusion with Fog. S2R+ cells adopt a flattened, discoid morphology when plated on concanavalin A (ConA)–treated coverslips. Within 10 min of Fog treatment the cells adopted a “puckered” shape and pushed their nuclei and organelles up and away from the coverslip. At the same time, radial, phase-dark furrows appeared at the cell periphery and moved centripetally to the center of the cell (Figure 1B). One of the downstream effects of Rho pathway signaling is activation of nonmuscle myosin II by phosphorylation of the motor's regulatory light chain (RLC). Therefore we treated S2R+ cells with concentrated Fog or control cell medium and examined the RLC phosphorylation state using phospho-specific antibodies. Immunofluorescence with phosphorylated-RLC antibodies revealed an overall increase in phosphorylation, along with a dramatic incorporation of myosin II into actomyosin purse-string structures (Figure 1C). To verify that Fog was acting via the canonical pathway involved in gastrulation, we used RNAi to deplete Cta, RhoGEF2, or Rho from S2R+ cells before Fog treatment. RNAi targeting Cta, RhoGEF2, or Rho prevented cellular constriction after Fog treatment (Figure 1, B and D), as did pretreatment of S2R+ cells with the Rho kinase small-molecule inhibitor Y-27632 (unpublished data). Previous work revealed that embryos mutant for the β subunit, β13F, and the γ subunit, γ1, exhibited gastrulation phenotypes similar to Cta mutants (Wang et al., 2005). We introduced RNAi targeted to these subunits and predicted that they comprise the heterodimer that associates with Cta. We found that these treatments blocked Fog-mediated contractility (Figure 1D). Thus we conclude that treatment of S2R+ cells with Fog activates the identical signaling pathway used in cellular contraction during Drosophila gastrulation.
Ric-8 is necessary for Fog pathway activation in Drosophila S2R+ cells
Next we tested the hypothesis that Ric-8 acts in the Fog pathway. We designed double-stranded RNAs (dsRNAs) to target the coding region or the 5′ and 3′ untranslated regions (UTRs) of the Ric-8 mRNA and found that each effectively depleted Ric-8 from S2R+ cells (Supplemental Figure S1). When tested in the contractility assay, Ric-8–depleted cells were unable to contract after treatment with Fog (Figure 1B and D). The effect of our RNAi was specific, as we were able to rescue the ability of S2R+ cells to respond to Fog by expressing Ric-8–green fluorescent protein (GFP) in cells depleted of endogenous Ric-8 (Figure 2, A and B). Ectopic overexpression of Ric-8–GFP was not sufficient, however, to induce contractility in the absence of Fog (unpublished data). From these data, we conclude that Ric-8 is a necessary component of the Fog signaling cascade.
FIGURE 2:
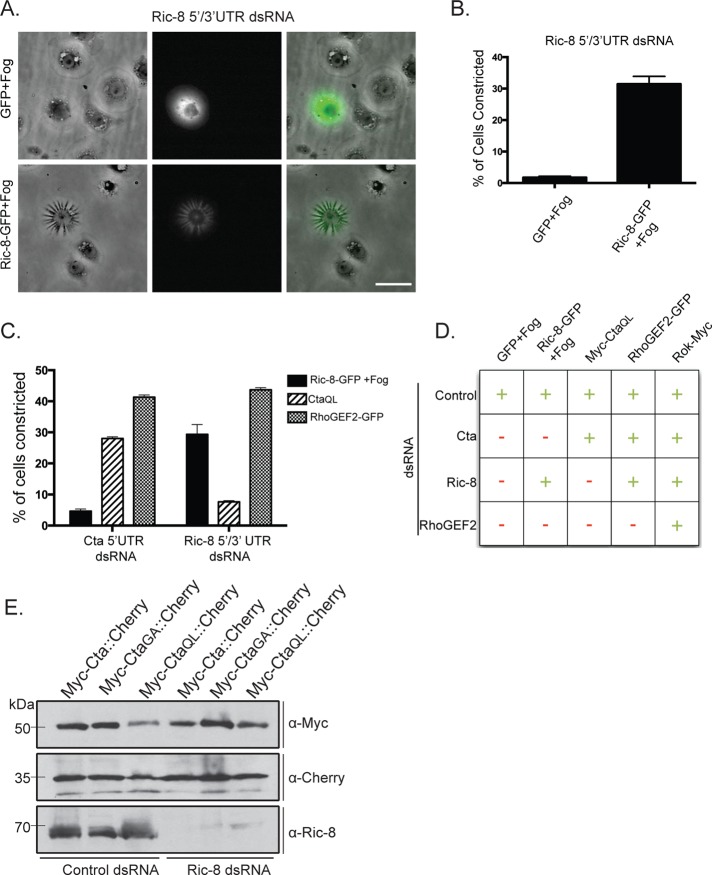
Ric-8 regulates the function of Cta within the Fog signaling pathway. (A) Expression of Ric-8-GFP, but not GFP alone, rescues the ability of cells depleted of endogenous Ric-8 to respond to Fog. Scale bar, 20 μm. (B) The number of GFP or Ric-8-GFP–transfected cells within a population depleted of endogenous Ric-8 was scored for their ability to contract in response to Fog (±SEM). (C) Cells depleted of endogenous Ric-8 or Cta were transfected with constitutively active Cta (CtaQL), RhoGEF2-GFP, or Ric-8-GFP + Fog treatment. Their ability to drive constriction was quantified as a percentage of the number of cells contracting within the population (±SEM). (D) Summary chart illustrating the epistatic relationship of Ric-8 in the Fog pathway. Transfected DNA and targeted dsRNA are indicated. +, ≥15% of transfected cells within a population constricted; –, ≤15% of transfected cells within a population constricted. (E) Cells were treated with control or Ric-8 dsRNA and transfected with a dual-expression construct for both Cta (WT, constitutively inactive:GA, or constitutively active:QL) and mCherry under separate promoters. Immunoblotting revealed equal amounts of Cta in control and Ric-8 dsRNA–treated cells; anti-dsRed was used as a protein loading control, and anti-Ric-8 to verify protein depletion.
To identify where Ric-8 is functioning within the Fog signaling pathway, we performed a series of epistasis experiments using RNAi. Overexpression of Myc-Cta-Q303L (CtaQL), a mutation predicted to lock Cta in its GTP-bound conformation (Xu et al., 1993), in S2R+ cells is sufficient to trigger contractility in the absence of Fog (see later discussion of Figure 4A). However, expression of Myc-CtaQL in S2R+ cells depleted of endogenous Ric-8 does not drive cellular constriction (Figure 2, C and D). To verify that the inability of Myc-CtaQL to trigger constriction in Ric-8–depleted cells was not due to the absence of overall Cta protein, we created dual-expression constructs with two distinct metallothionein promoters within the same vector: 1) preceding full-length Myc-Cta and 2) preceding the coding sequence for mCherry. We then transfected these constructs into S2 cells treated with either control or Ric-8 dsRNA and compared levels of wild-type Myc-Cta, constitutively active Myc-CtaQL, and constitutively inactive Myc-Cta-G302A (CtaGA). Mutation of glycine 302 to alanine is predicted to trap Cta in either its GDP-bound conformation or in a nucleotide-free state, based on homology with similar mutations in other Gα12/13 family members (Gohla et al., 1999). Using these constructs, we show that levels of ectopic Myc-Cta are not affected by Ric-8 depletion (Figure 2E). In the converse experiment, S2R+ cells depleted of endogenous Cta and overexpressing Ric-8–GFP do not constrict upon Fog treatment. However, overexpression of RhoGEF2, which is directly downstream of Cta, in either Cta- or Ric-8–depleted cells is sufficient for cellular constriction (Figure 2, C and D). Therefore Ric-8 functions upstream of RhoGEF2, implying a role for Ric-8 at the level of the putative GPCR, Cta, or the βγ subunits. It has been well documented in Drosophila and other systems that Ric-8 does not interact with Gα when it is complexed with its βγ subunits (Tall et al., 2003; Hampoelz et al., 2005; Wang et al., 2005). There is no evidence that Ric-8 interacts with a receptor in receptor-dependent activation of Ric-8, although this possibility has not been directly tested.
FIGURE 4:
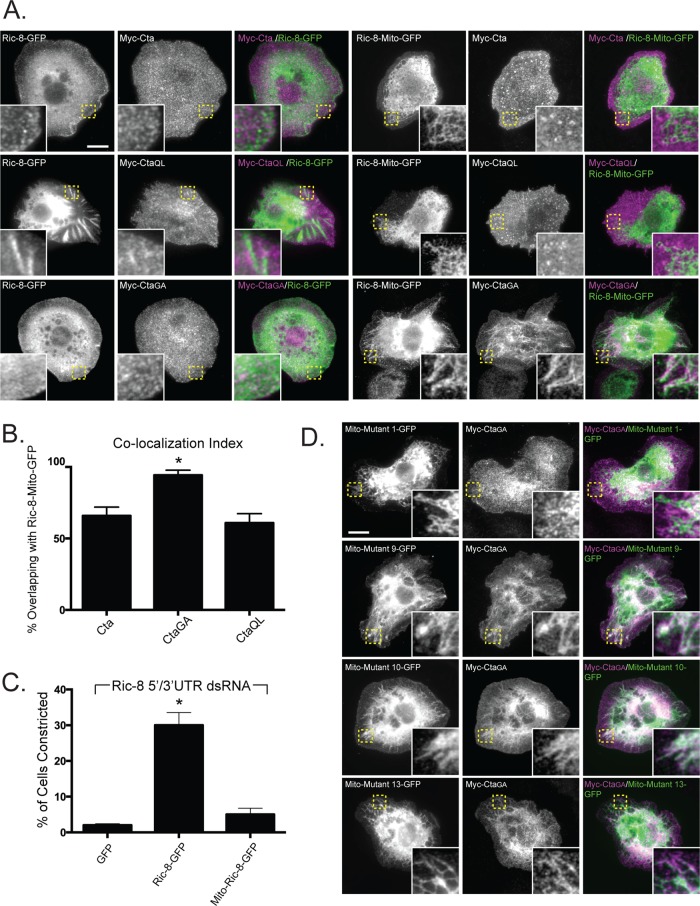
Ectopic localization of Ric-8 drives mislocalization of constitutively inactive Cta and attenuates the efficacy of Fog signaling. (A) Targeting Ric-8 to the mitochondria causes CtaGA but not Myc-Cta or Myc-CtaQL to localize to the mitochondria. S2 cells were transfected with Ric-8–GFP, or Mito-Ric-8–GFP, and Myc-Cta, Myc-CtaGA, or Myc-CtaQL and stained for anti-Myc and anti-GFP. Insets, enlarged views of boxed images in Mito-Ric-8–GFP and Cta variants. Scale bar, 20 μm. (B) Colocalization of Mito-Ric-8–GFP/Cta is greater with inactive Cta than with wild-type or constitutively active Cta. Colocalization index was determined using the Manders coefficient (the overlap of Concertina with Ric-8-Mito–GFP; ±SEM; error bars, p < 0.05). (C) Mislocalized Ric-8 fails to compensate to drive Fog-induced cellular constriction. S2R+ cells depleted of endogenous Ric-8 transfected with GFP, Ric-8-GFP, and Mito-Ric-8–GFP were treated with Fog and scored for their ability to constrict (±SEM; error bars, p < 0.05). (D) Evolutionarily conserved residues contribute to localization of CtaGA. Mito-tagged versions of Ric-8–GFP mutants that exhibit decreased Myc-CtaGA binding (Figure 5, B and C) were coexpressed with Myc-CtaGA. Whereas mutants 9, 10, and 13 colocalized with Myc-CtaGA, mutant 1 did not. Insets, enlarged images of boxed areas. Scale bar, 20 μm.
Ric-8 directly binds Cta and exhibits higher affinity for the inactive form of Cta
Given the likelihood that Ric-8 interacts with the Gα in our system, we used immunoprecipitation to test the hypothesis that Ric-8 and Cta directly interact. A disadvantage of this strategy is that antibodies against Cta have not been published and our attempts to develop them were unsuccessful. However, we found that Myc-Cta is functional and able to restore Fog sensitivity to S2R+ cells depleted of endogenous Cta by RNAi (Supplemental Figure S2, A and B); thus we used this construct as a proxy for endogenous protein. We transfected Myc-Cta into S2 cells and immunoprecipitated with an anti-Myc monoclonal antibody, and found that endogenous Ric-8 coprecipitated (Figure 3A). As expected from our rescue experiments, Ric-8–GFP also coimmunoprecipitated with Myc-Cta (Figure 3B). Thus, Ric-8 and Cta are able to interact in Drosophila tissue culture cells.
FIGURE 3:
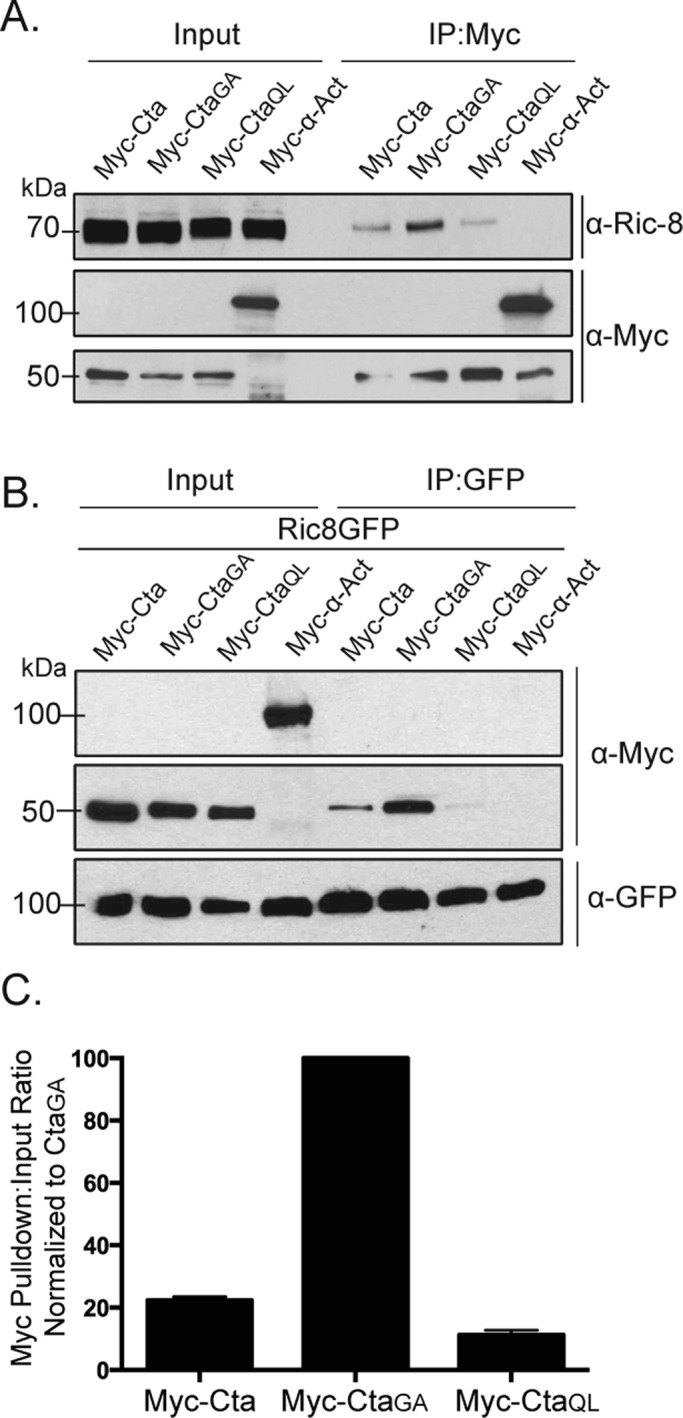
Ric-8 physically interacts with Cta and exhibits higher binding affinity for constitutively inactive Cta. (A) S2 cells were transfected with Myc-Cta, Myc-CtaGA, Myc-CtaQL, or α-actinin (α-Act) as a negative control. IPs were performed using an anti-Myc antibody, and samples were probed with anti–Ric-8 and anti-Myc. (B) S2 cells were transfected with Ric-8–GFP and Myc-Cta, Myc-CtaGA, Myc-CtaQL, or α-Act. IPs were performed with GFP-binding protein and probed with anti-GFP and anti-Myc. (C) Quantification of IPs performed in B. Pull-down:input ratios were determined using quantitative densitometry and normalized against CtaGA (±SEM).
Given that Ric-8 functions as a GEF for Gα subunits in other systems, we wanted to test the hypothesis that Ric-8 exhibits a preferred interaction with GTP-free Cta. To do this, we used an inactive version of Cta, CtaGA. CtaGA fails to rescue the contractility effects of S2R+ cells depleted of endogenous Cta (Supplemental Figure S2B). To determine whether nucleotide association affected Ric-8 interaction, we cotransfected Ric-8–GFP together with wild-type Myc-Cta, Myc-CtaQL, or Myc-CtaGA into S2 cells. We prepared lysates from transfected cultures, immunoprecipitated GFP, and compared the amount of Myc-Cta in each sample by quantitative immunoblot. We found that Ric-8 binding to Cta is dependent on the nucleotide state of Cta, as pull-downs performed with constitutively inactive Myc-CtaGA and constitutively active Myc-CtaQL showed greater and lesser binding affinity to Ric-8, respectively, than wild-type Myc-Cta (Figure 3, B and C). These data indicate that Ric-8 discriminates between Cta nucleotide states and preferentially binds to inactive Cta.
Ric-8 acts to selectively localize nucleotide-free Cta within the cell
Ric-8 is necessary to localize Cta and Gαi to the cortex in Drosophila embryonic epithelium (David et al., 2005; Hampoelz et al., 2005; Wang et al., 2005; Kanesaki et al., 2013). In S2 cells, Myc-Cta normally localizes to the plasma membrane and exhibits enrichment in membrane ruffles. On Ric-8 depletion, we saw a reduction of Cta at the plasma membrane and an absence in ruffles (Supplemental Figure S3). We therefore wanted to test the hypothesis that Ric-8 functions to localize Cta and determine whether its nucleotide state plays a role in this interaction. Our strategy was to coexpress Myc-Cta along with a version of Ric-8-GFP that was mistargeted to mitochondria by tagging it with residues 310–338 of Listeria ActA (mitochondrially tagged [Mito]-Ric-8–GFP). We expressed Mito-Ric-8–GFP in S2 cells and showed that it localized to the mitochondria (Supplemental Figure S4). When wild-type Myc-Cta or Myc-CtaQL was coexpressed with Mito-Ric-8–GFP, neither Cta construct exhibited discrete localization (Figure 4, A and B). However, coexpression of Myc-CtaGA and Mito-Ric-8–GFP resulted in robust accumulation of Cta to the mitochondria (Figure 4, A and B). These data indicate that Ric-8 acts to selectively localize inactive Cta within the cell.
Our results suggest Ric-8 could act to localize Cta to the cell cortex to mediate Fog signaling. To test this model, we transfected S2R+ cells depleted of endogenous Ric-8 with either Ric-8–GFP or Mito-Ric-8–GFP and scored for the ability of each construct to rescue contractility. Ric-8–GFP restored the normal constriction of Ric-8 depleted cells; however, cells expressing Mito-Ric-8–GFP exhibited a significantly diminished response to Fog (Figure 4C). These results suggest that either Ric-8 needs to be cytoplasmic to function correctly or the Mito-Ric–8 is sequestering endogenous Cta to the mitochondria.
Ric-8 binds to Cta through an interface of conserved residues
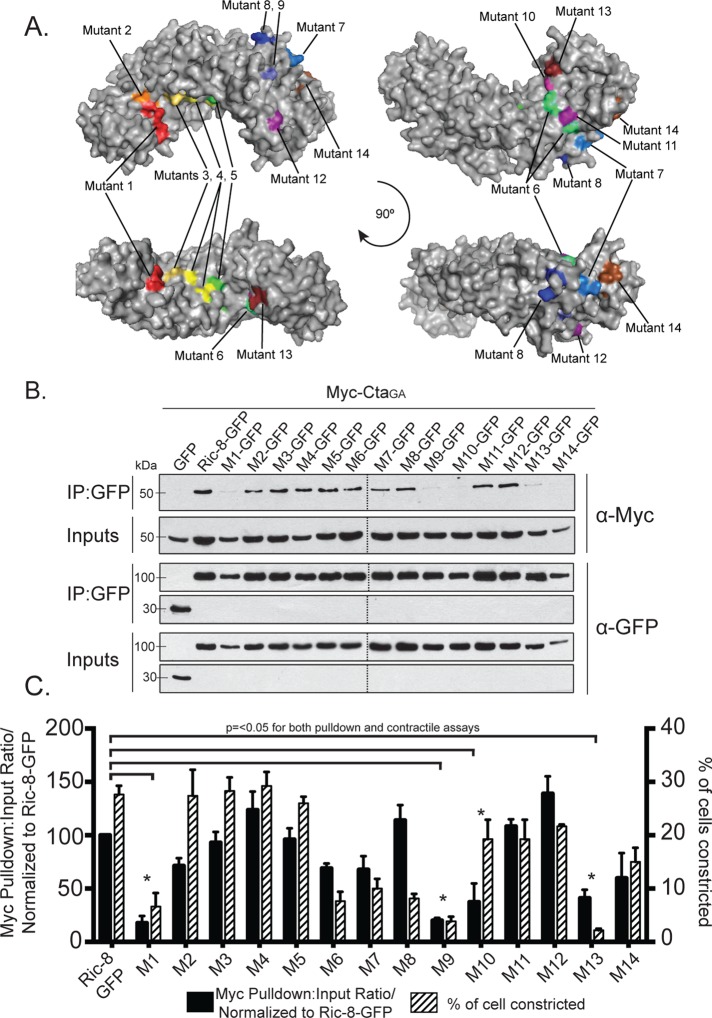
Although previous work provided insight into the structure of Ric-8, a rigorous investigation of specific residues important for interactions with Gα has not been performed. Ric-8 is predicted to be composed of 10 Armadillo repeats (Figueroa et al., 2009). Armadillo repeats adhere to a canonical fold and global elongated structure (Coates, 2003). The Olate group recently used molecular modeling to construct an in silico model of the Ric-8 structure (Figueroa et al., 2009). On the basis of sequence conservation of Ric-8 across species (Supplemental Figure S5), we made 14 cluster mutations in Ric-8–GFP, targeting conserved electrostatic residues likely to be surface exposed and that were exposed in the Ric-8 model (Table 1 and Figure 5A). These mutations consisted of charge reversals, with the intent not only to diminish, but also to repel an interaction with Cta. We coexpressed the Ric-8–GFP mutants with Myc-Cta, Myc-CtaQL, or Myc-CtaGA in S2 cells and assessed their ability to interact. Several of our Ric-8–GFP mutant constructs exhibited altered affinities for Cta and are described later. The mutants span the length of the protein and are ordered in succession from N-terminus to C-terminus. Although Ric-8–GFP robustly bound Myc-CtaGA, it exhibited lower-affinity interactions with Myc-Cta and Myc-CtaQL (Figure 2, B and C). Owing to this weaker binding, Ric-8–GFP pull-down experiments performed with either Myc-Cta or Myc-CtaQL displayed a high degree of variance, making it difficult to determine interaction strengths between these molecules. Therefore we focused our analyses on Ric-8–GFP and Myc-CtaGA.
TABLE 1:
Cluster mutations used in Ric-8 binding and Fog-induced cellular contractility assays.
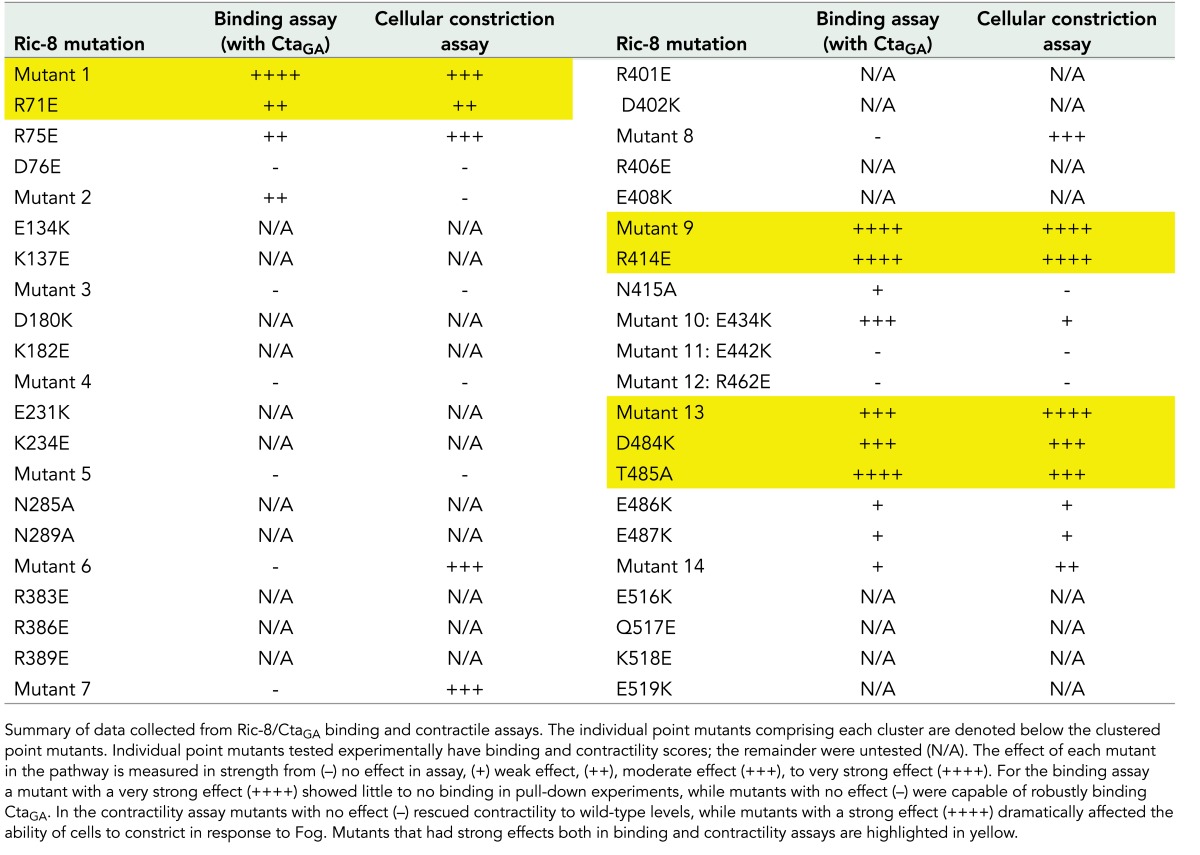
FIGURE 5:
Evolutionarily conserved electrostatic residues are required for binding between Ric-8 and CtaGA. (A) Clusters of point mutants used in our screen are represented by different colors on a model of Ric-8. (B, C) Mutant clusters within Ric-8 disrupt binding to CtaGA and inhibit pathway activation downstream of Fog. (B) S2 cells were transfected with GFP, Ric-8–GFP, or cluster Ric-8–GFP mutants and CtaGA. IPs were performed with GFP-binding protein and probed with anti-GFP and anti-Myc. (C) The pull-down:input ratios for Ric-8–GFP and Myc-CtaGA were quantified using densitometry and normalized to Ric-8–GFP (±SEM; error bars, p < 0.05; black bars). S2R+ cells were depleted of endogenous Ric-8 and transfected with Ric-8–GFP and cluster Ric-8–GFP mutants and then scored for the percentage of transfected cells constricting within the population (±SEM; error bars, p < 0.05; hatched bars). Dashed lines indicate where two separate gels have been combined.
We identified four Ric-8–GFP mutants with clustered point mutations (1, 9, 10, and 13) that had significantly reduced binding to Myc-CtaGA (Figure 5, B and C, and Table 1). To further parse out the individual residues responsible for this interaction, we made single point mutants for each cluster of more than one mutated amino acid (Supplemental Figure S6 and Table 1). Mutant 10 is a singular mutation, so it was not tested again. We identified specific residues within mutants 1, 9, and 13 that attenuated the ability of Cta to bind Ric-8 (Figure 6, A and B, and Table 1). Although not statistically significant, both Myc-Cta and Myc-CtaQL variants exhibited decreased binding to mutant 1 and moderate to high binding to mutants 9, 10, and 13 (Supplemental Figure S7, A–D). The fact that mutants 9, 10, and 13 inhibited binding to Myc-CtaGA but did not strongly affect Myc-Cta or Myc-CtaQL binding indicates that the C-terminal region of Ric-8 may be important for high-affinity binding seen specifically in the Ric-8/GTP–free Cta interaction, whereas the N-terminal residues found in mutant 1 could be important for global Ric-8 association and function.
FIGURE 6:
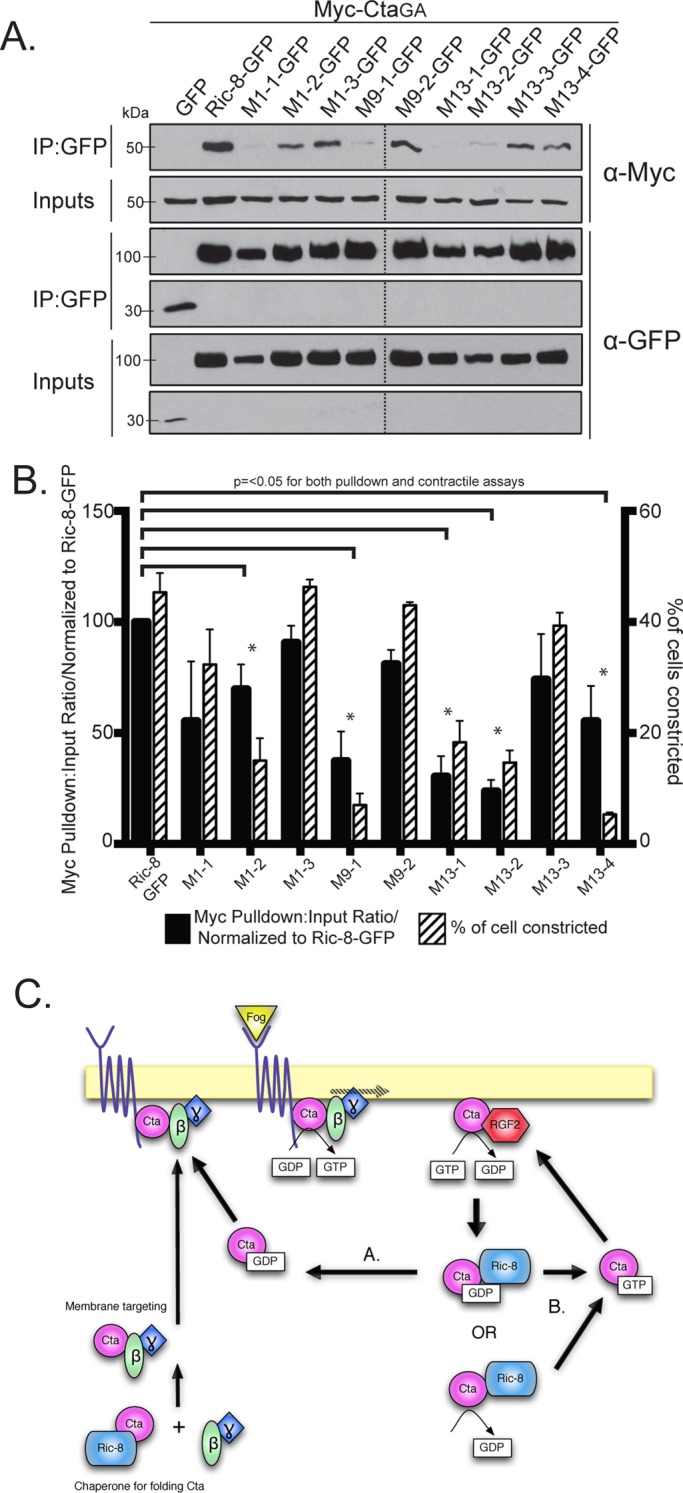
Individual residues derived from Ric-8 cluster mutants comprise key interaction sites for Cta binding and function. (A) Individual Ric-8 point mutants from cluster mutants (1, 9, and 13) negatively regulate binding to Myc-CtaGA. Cells were transfected with GFP, Ric-8–GFP, or individual Ric-8–GFP point mutants and CtaGA. IPs were performed with GFP-binding protein and probed with anti-GFP and anti-Myc. (B) Quantification of the IP experiments in A (black bars). The pull-down:input ratios were determined using quantitative densitometry and normalized to Ric-8–GFP (±SEM; error bars, p < 0.05). S2R+ cells were depleted of endogenous Ric-8, transfected with Ric-8–GFP or individual Ric-8–GFP point mutants, and scored for the percentage of transfected cells constricting within the population (±SEM; hatched bars). Dashed lines indicate where two separate gels have been combined. (C) Proposed model for Ric-8 function within the Fog signaling pathway. Ric-8 initially acts to chaperone the folding of Cta and is released before Cta association with β13F and γ1. The heterotrimer is targeted to the plasma membrane, where it interacts with a GPCR for Fog. Fog binding activates Cta through exchange of GDP for GTP. Cta-GTP activates RhoGEF2, and RhoGEF2’s GAP activity catalyzes hydrolysis of GTP to GDP. Ric-8 may then either bind Cta-GDP or stabilize a nucleotide-free version of Cta. Ric-8 then localizes the inactive Cta for reactivation and reinsertion into the Fog signaling pathway, either by returning it to the heterotrimer to be reactivated by the GPCR (A) or through disassociation of GDP, which facilitates GTP binding, and subsequent pathway reinsertion directly upstream of RhoGEF2 (B).
We next tested the hypothesis that the residues mediating Ric-8/Cta interactions are required for Fog signaling. We depleted endogenous Ric-8 from S2R+ cells and transfected the cells with the clustered and individual point mutant variants of Ric-8-GFP. We then treated the cells with Fog and assessed the ability of the transfected cells to rescue constriction. Of the 14 clustered point mutants tested in the binding assay, six (1, 6–9, and 13) failed to rescue Fog-induced constriction (Figure 5C and Table 1). Testing the individual point mutants within the cellular constriction assay revealed a similar pattern in the residues that prevented pathway activation to the individual residues deficient in binding Cta in the pull-down assay (Figure 6, A and B, and Table 1). This suggests that residues R71, R75, R414, D484, T485, and E487 within Ric-8 act to establish a binding interface with Cta and are important for successful G protein signaling downstream of Fog.
To determine whether Ric-8 mutants with low Cta-binding affinity affected Cta localization, we made Mito-tagged versions of cluster mutants 1, 9, 10, and 13. We transfected S2 cells with the Mito-Ric-8–GFP cluster mutants and Myc-CtaGA and screened for colocalization of the two proteins. Myc-CtaGA did not colocalize with Mito-Ric-8–GFP mutant 1, whereas, surprisingly, colocalization of Myc-CtaGA was seen with Mito-Ric-8–GFP mutants 9, 10, and 13 (Figure 4D). Hypothetically, mutants 9, 10, and 13 could be affecting the binding kinetics of Myc-CtaGA, which may account for the colocalization of the two in our mistargeting assays, as well as the absence of interaction within the pull-down assay.
Mutants 1, 9, and 13 had low binding affinity to CtaGA, as well as decreased contractility in Fog-treated S2R+ cells; however, mutant 10 rescues contractility to near-wild-type levels (Figure 5, B and C, and Table 1). It is possible that mutant 10, while impeding the binding interface between Ric-8 and Cta, is sufficient in its interaction to function in pathway activation. Intriguingly, mutants 6–8 are capable of binding Cta, as shown in pull-down assays (Figure 5, B and C, and Table 1) but have diminished ability in activating the pathway (Figure 5C and Table 1), suggesting that these residues may have an important functional role outside of binding. The majority of the residues that affected both binding and Fog-induced contraction rescue map to a conserved face of the Arm repeats and show a potential clamp-like binding of Ric-8 to Cta (Supplemental Figure S6). Our findings suggest that residues within the inner face of the N-terminus (R71 and R75) of Ric-8 may act to facilitate global interaction with Cta, whereas residues found in the C-terminus (R414, D484, T485, and E487) modulate binding based on nucleotide specificity. Whereas previous work found residues important in Gα subunits for facilitating interaction with Ric-8 (Thomas et al., 2011), here we identify some of the first residues found to be important for Ric-8 binding to a Gα.
DISCUSSION
We established a novel assay for testing potential Fog pathway components and found that in Drosophila tissue culture Ric-8 is required for pathway activation and not only binds the Gα12/13, Cta, but also preferentially binds inactive Cta, CtaGA. We defined a role for Ric-8 as an escort/scaffold for CtaGA by using artificially induced localization of Ric-8 to the mitochondria. On Ric-8 translocation, we found that CtaGA colocalized with ectopically localized Ric-8, whereas the cellular localization of wild-type and constitutively active Cta was unaffected. In addition, when Ric-8 was mistargeted to the mitochondria, cells were impaired in their ability to constrict in response to Fog application. Further, we identified evolutionarily conserved residues within Ric-8 potentially important for 1) establishment of a Ric-8/Cta–binding interface, 2) nucleotide-specific recognition of Cta, and 3) successful G protein signaling downstream of Fog.
Our novel cell-based assay is ideal for examining Fog-induced activation of the Rho pathway due to the ease with which we are able to deplete cells of specific proteins using RNAi, the rapidity of screening multiple genes simultaneously, and the ability to visualize pathway activation using simple microscope-based examination. This assay opens numerous possibilities for the identification of other pathway components, including the unidentified GPCR involved in transduction of the Fog signal, as well as investigation of general cellular functions such as mechanochemical force production and regulation of the actomyosin cytoskeleton. In addition, although not highlighted in this study, we are able to view Fog-induced contractility in real time. This allows for further investigation of pathway components that specifically affect the kinetics of Fog responsiveness and/or the longevity and persistence of pathway activation. In Drosophila, and other systems, Ric-8 modulates the behavior of Gα subunits during asymmetric cell divisions (reviewed in Hinrichs et al., 2012). Owing to its role in establishing asymmetry in dividing cells and subsequently controlling cell proliferation rates, Ric-8 has become of interest in the field of cancer biology (Luo et al., 2008; Muggerud et al., 2009). Our model cell culture system provides a streamlined approach for further investigation into parsing out the complicated signaling networks involved in establishing these disease states.
Previous work implicated Ric-8 as a chaperone during Gα biosynthesis to stabilize nascent protein production and in turn as an essential factor in Gα membrane targeting. This function of Ric-8 affects the stability of all classes of mammalian Gα subunits (Gabay et al., 2011; Thomas et al., 2011). Given the necessity of Ric-8 in mammalian systems for Gα stabilization and membrane localization, it is likely that Ric-8 acts similarly in Drosophila, as evidenced by the mistargeting of Gαi and Cta, in the absence of Ric-8, to the cortex of the epithelium of Drosophila embryos (David et al., 2005; Hampoelz et al., 2005; Wang et al., 2005; Kanesaki et al., 2013) and the mislocalization of Cta in Drosophila tissue culture cells (Supplemental Figure S3). However, unlike Gαi (Hampoelz et al., 2005), the levels of Cta are not dramatically affected in the absence of Ric-8 (Figure 2E); in addition, we see some rescue in cells depleted of endogenous Ric-8, overexpressing constitutively active Cta (Figure 2C), indicating that at least a small amount of Cta is localized correctly and functional. Therefore, whereas plasma membrane levels of Cta are affected by Ric-8 (Kanesaki et al., 2013; present work), overall levels of protein are not (Figure 2E). One possibility, given that constitutively active Cta was still able to rescue, is that Ric-8 could be important for Gα cycling at the site of receptor activation, which is believed important for spatial regulation of Gα signaling (Ross, 2008).
Although signaling nodes involving GPCRs, Gα subunits, and Ric-8 have been extensively studied, little is known about the structure of Ric-8 and how it interacts with Gα. We used a predicted model (Figueroa et al., 2009) of Ric-8 as a conceptual basis to visualize mutants and identify key conserved residues important for Cta binding, nucleotide specificity, and execution of productive G protein pathway activation. Based on these data, our structure–function assay of Ric-8 identified four cluster mutations (1, 9, 10, and 13; Supplemental Figure S6) that inhibited CtaGA binding, of which three (1, 9, and 13) also failed to rescue Fog-induced constriction to wild-type levels. Of these four mutants, we found that only mutant 1 (in the N-terminus of Ric-8) had an inhibitory effect on binding to wild-type, constitutively active, and constitutively inactive Cta, whereas mutants 9, 10, and 13 (in the C-terminus of Ric-8) were deficient only in binding inactive Cta. The Itoh lab found that a truncated version consisting of the N-terminal half (residues 1–301) of Ric-8 was sufficient to bind Gαq (Nishimura et al., 2006). In accordance with these data, we suggest that residues in mutant 1 are important for non–nucleotide-specific Cta interaction, whereas residues in mutants 9, 10, and 13 confer nucleotide-specific recognition of Cta. This study presents the first evidence of specific residues within Ric-8 facilitating interaction with a Gα.
Several mutants had effects in only the binding or contractile assay. Mutant 10 inhibited binding, whereas mutants 6–8 prevented Fog-induced constriction. Mutant 10 was able to modestly rescue cellular constriction but exhibited decreased binding to Cta, implying that this mutant is still functional but perhaps folded in a manner unproductive for robust binding to Cta; this may be due to its proximity to mutant 13 (Figure 5A). Mutants 6–8 are capable of binding Cta but not rescuing Ric-8 function downstream of pathway activation. Although the function of mutant clusters 6–8 is unclear, it is tempting to hypothesize that this region is a potential site for Ric-8 GEF activity.
In the early dividing C. elegans embryo (Afshar et al., 2005), D. melanogaster neuroblasts and epithelium (David et al., 2005; Hampoelz et al., 2005; Wang et al., 2005; Kanesaki et al., 2013), and several mammalian tissue culture cell lines (Woodard et al., 2010; Gabay et al., 2011; Wang et al., 2011) Ric-8 localizes Gα subunits to the plasma membrane. Our data suggest that there is an additional level regulating Gα localization that is dependent on the nucleotide-bound state of Gα. We identified a cluster of residues that may facilitate this interaction with Cta. Clustered Ric-8 mutants, deficient in binding CtaGA in immunoprecipitation assays, when tagged with a sequence directing them to the mitochondria had varying effects in their ability to ectopically localize CtaGA. Mito-Ric-8 mutant 1 did not recruit CtaGA to its ectopic location at the mitochondria, whereas Mito-Ric-8 mutants 9, 10, and 13 triggered mitochondrial mislocalization of CtaGA (Figure 4D). Of interest, mutants 9, 10, and 13 exhibited decreased binding to constitutively inactive Cta, CtaGA, but not to wild-type or constitutively active Cta, CtaQL (Figure 5, B and C, and Supplemental Figure S7). This implies that these residues may confer temporally regulated nucleotide specific recognition sites for Cta.
On the basis of our characterization of Ric-8 and data from the literature, we propose the following model (Figure 6C). Ric-8 acts to initially chaperone the folding of Cta, allowing Cta, Gβ13F, and Gγ1 to form a complex that is then transported to the plasma membrane (Gabay et al., 2011; Kanesaki et al., 2013). On Fog/GPCR interaction, GTP-bound Cta is released from the Gβγ heterodimer and interacts with RhoGEF2 (via its RGS domain), causing hydrolysis of GTP to GDP. Specific, evolutionarily conserved residues regulate the binding of GDP-bound Cta to Ric-8, or, alternatively, Ric-8 stabilizes a nucleotide-free version of Cta. This allows Cta to bypass destruction and be reinserted into the Fog pathway to activate downstream targets.
MATERIALS AND METHODS
Tissue culture, transfection, and RNAi
S2 and S2R+ cell lines were obtained from the Drosophila Genome Resource Center (Bloomington, IL), and propagated as previously described (Rogers and Rogers, 2008). S2 cells were maintained in SF900 SFM (Invitrogen, Carlsbad, CA) and S2R+ cells in Shields and Sang medium (Invitrogen) supplemented with 5% heat-inactivated fetal bovine serum (Invitrogen). S2 and S2R+ cells were transfected with 2 μg/μl DNA using the Amaxa Nucleofector system with Kit V using program G-30 (Lonza, Basel, Switzerland), or with FuGENE HD (Promega, Madison, WI; except the Mito-tag constructs, for which 1 μg/μl DNA was used). For individual RNAi treatments, cells at 75–90% confluency in 6- or 12-well plates were treated every other day for at least 10 d with 15 μg/ml dsRNA. dsRNAs were produced using Promega Ribomax T7 kit according to instructions. Primers used for dsRNA synthesis are as follows, with all preceded by the T7 sequence (5′-TAATACGACTCACTATAGG-3′). Control, forward (fwd), 5′-TAAATTGTAAGCGTTAATATTTTG-3′, and Control, reverse (rev), 5′-AATTCGATATCAAGCTTATCGAT-3′, to amplify a region from the pBluescript plasmid; Cta, fwd, 5′-TGACCAAATTAACTCAAGAACGAAT-3′, and Cta, rev, 5′- TTCCAGGAACTTATCAATCTCTTTG-3′; Cta 5′ UTR, fwd, 5′-ATATACAGGCAAAAATTATTATCACCGCTGTTGTTTGC-3′, and Cta 5′ UTR, rev, 5′-CGCTGGCAAGCCAACGCCTGATGCTCGCACTTTCTATA-3′; RhoGEF2, fwd, 5′-ATGGATCACCCATCAATCAAAAAACGG-3′, and RhoGEF2, rev, 5′- TGTCCCGATCCCTATGACCACTAAGGC-3′; Rho, fwd, 5′-GTAAAACTTGCCTTCTGATTGTCT-3′, and Rho, rev, 3′-ATCTGGTCTTCTTCCTCTTTTTGA-3′; Ric-8, fwd, 5′-GCAGGCGCCAGTGCCTGCGGC-3′, and Ric-8, rev, 5′-CCGGAGATGTTTGTCAGCA-3′; Ric-8 5′ UTR, fwd, 5′-GCAAAGGTGCGGTCAC-3′, and Ric-8 5′ UTR, rev, 5′-GTCGCCAACGGTGGC-3′; and Ric-8 3′ UTR, fwd, 5′-GTATTGCGGGATCTG-3′, and Ric-8 3′ UTR, rev, 5′-GGGCGTGTATTTAA-3′.
Contractility assay
S2R+ cells were resuspended and plated on concanavalin A (MP Biomedicals, Solon, OH)–coated coverslips, allowed to spread for 1–3 h, and then treated for 10 min with concentrated Fog-conditioned medium or medium harvested from S2 cells. To produce Fog-conditioned medium, we created a stable S2 cell line carrying a Fog-Myc expression construct driven by an inducible metallothionein promoter. Fog-stable S2 cells were grown to 75–90% confluency in T150 flasks before SF-900 media was exchanged for Schneider's media (Invitrogen) and induced with 1 mM CuSO4 for 48 h. Cells were then pelleted, and the supernatant was concentrated using protein concentrators (Millipore, Billerica, MA) to ∼2.5–5% of the original volume. For control media, the same process was applied to non–Fog-expressing S2 cells. Control (S2) and Fog-concentrated media were diluted 1:1 with Schneider's media before application. For each experiment, we scored the number of cells within a population that contracted in response to Fog treatment, repeating each condition at least three times, and counted ≥500 cells. Error bars were calculated using SE.
Molecular biology
The Fog-Myc expression construct was generated using PCR to amplify the coding sequence of the gene and introduce a 5′ EcoRI site, a C-terminal Myc tag, and a 3′ NotI site to allow cloning into pMT-V5/His (Invitrogen). Construction of N-terminally Myc-tagged Cta and C-terminally GFP-tagged RhoGEF2 constructs was described previously (Rogers et al., 2004). The dual-expression constructs were created by subcloning Myc-Cta constructs into pMT-V5-histidine (His) containing a second transcriptional unit for membrane-mCherry marker containing the sqh promoter and 3′ UTR (Martin et al., 2010). To generate the expression construct for constitutively active Rho kinase, we used PCR to amplify the catalytic domain (amino acids 1–506) from a cDNA (expressed sequence tag clone LD15203) and introduced a 5′ EcoRI site and a 3′ NotI site and incorporated the Myc epitope tag at the 5′ end of the coding sequence. This insert was then subcloned into pMT-A for inducible expression. Full-length Ric-8a cDNA was subcloned using the Gateway TopoD pEntr system (Invitrogen) into a final Zeocin-selectable pIZ backbone that has a metallothionein promoter, Gateway (Invitrogen) LR recombination sites in the multiple cloning site, and a C-terminal enhanced GFP tag. All mutagenesis was preformed on this construct using KOD Xtreme Hot Start Polymerase (Novagen, Gibbstown, NJ). Mitochondrial localization of Ric-8 was achieved by N-terminally attaching Listeria monocytogenes ActA residues 310–338 (Pistor et al., 1994).
Immunoprecipitation and immunoblotting
We bacterially expressed a His- and Fc-tagged GFP-binding protein (Fc-GFP-BP). The Fc-GFP-BP was first purified on a Ni column, and the eluted Fc-GFP-BP fractions were incubated with protein A beads. GFP-binding- protein was covalently linked to the beads using 20 mM dimethylpimelimidate (Sigma-Aldrich, St. Louis, MO). Before use in immunoprecipitation (IP) experiments, beads were washed with IP lysis buffer (50 mM Tris, 150 mM NaCl, 0.5 mM EDTA, 1 mM dithiothreitol, 0.5% Triton X-100, 2.5 mM phenylmethylsulfonyl fluoride, and Complete EDTA-Free Protease Inhibitor Cocktail [Roche, Indianapolis, IN]).
S2 cells used for IPs were transfected as described and induced 24 h later with 1 mM CuSO4. The next day cells were resuspended, pelleted, and washed before lysing with IP lysis buffer. Samples were removed for input controls, and the rest of the sample was incubated with GFP-binding-protein beads. Samples were resuspended in SDS–PAGE sample buffer and boiled for 10 min. SDS–PAGE sample buffer was also added to input samples and boiled for 10 min. Samples were run on SDS–PAGE gels and transferred to nitrocellulose membranes for Western blotting using anti-Myc9e10 (Developmental Studies Hybridoma Bank [DSHB], Iowa City, IA) and anti-GFPJL8 (Clontech, Mountain View, CA). For immunoblot quantitation the pull-down:input ratios were determined using densitometry with ImageJ software (National Institutes of Health, Bethesda, MD) on scanned film images. All immunoblot quantitation was performed at least three times on three distinct blots. Error bars represent SEM. Asterisks in Figures 3, B and C, 5C, and 6B and Supplemental Figure S7B indicate p < 0.05 determined using Student's t test.
Microscopy
Cells were plated onto ConA-coated coverslips and prepared for imaging as previously described (Rogers and Rogers, 2008), differing only in the addition of 0.08% formic acid to the fixation solution of cells stained with the anti-ATP synthase α subunit antibody. Antibodies used for immunofluorescence were as follows: anti–phospho-myosin light chain 2 (Ser-19; Cell Signaling Technology, Danvers, MA), anti–Ric-8 (gift from William Chia, National University of Singapore), anti-Fog (gift from Eric Wieschaus, Princeton University), anti-dsRed (Clontech), anti-GFPJL8 (Clontech), anti-Myc9e10 (DSHB), anti–ATP synthase α subunit (MitoSciences, Eugene, OR), and anti-DM1α and Alexa Fluor 564–phalloidin (Invitrogen). All cells were imaged using a CoolSnap HQ charge-coupled device camera (Roper Scientific, Tucson, AZ) mounted on an Eclipse Ti-E and driven by Nikon Elements software (Nikon, Melville, NY), except cells in Figure 4, which were imaged using a total internal reflection fluorescence system (Nikon) mounted on an inverted Ti-E microscope using an Andor-Clara Interline camera (Andor Technology, Belfast, UK) and driven by Nikon Elements software, and cells in Supplemental Figure S3, which were imaged using an inverted IX81 (Olympus, Tokyo, Japan) with a CoolSnap ES camera (Photometrics, Tucson, AZ) driven by MetaMorph software (Molecular Devices, Sunnyvale, California). Colocalization index was determined using the Manders coefficient powered by the ImageJ plug-in JACoP (for more information see Bolte and Cordelières, 2006). Photoshop CS3 (Adobe, San Jose, CA) was used to adjust input levels so that the main range of signals spanned the entire output grayscale and to adjust brightness and contrast.
Supplementary Material
Acknowledgments
We thank T. Meigs, M. Peifer, K. Slep, and J. Sondek for thoughtful discussion and feedback on the manuscript. We thank D. Bosch and D. Siderovski for collaborative efforts to purify Drosophila Ric-8 and Cta, D. Marston for help with imaging, and J. Olate for the xRic-8 PDB file. We thank G. Rogers, W. Chia, and E. Wieschaus for reagents. This work was supported by grants from the National Institutes of Health (RO1-GM081645 to S.L.R.) and the Arnold and Mabel Beckman Foundation (Beckman Young Investigator Award to S.L.R.). K.A.P. was supported by funding from the Lineberger Comprehensive Cancer Center.
Abbreviations used:
- Cta
Concertina
- Fog
Folded gastrulation
- GDI
guanine-nucleotide dissociation inhibitor
- GEF
guanine nucleotide exchange factor
- GPCR
G protein–coupled receptor
- Mito
mitochondrially tagged
- S2R+
S2-receptor +
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0813) on September 4, 2013.
REFERENCES
- Afshar K, Willard FS, Colombo K, Johnston CA, McCudden CR, Siderovski DP, Gönczy P. RIC-8 is required for GPR-1/2-dependent Galpha function during asymmetric division of C. elegans embryos. Cell. 2004;119:219–230. doi: 10.1016/j.cell.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Afshar K, Willard FS, Colombo K, Siderovski DP, Gönczy P. Cortical localization of the Galpha protein GPA-16 requires RIC-8 function during C. elegans asymmetric cell division. Development. 2005;132:4449–4459. doi: 10.1242/dev.02039. [DOI] [PubMed] [Google Scholar]
- Barrett K, Leptin M, Settleman J. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 1997;91:905–915. doi: 10.1016/s0092-8674(00)80482-1. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Chan P, Gabay M, Wright FA, Tall GG. Ric-8B is a GTP-dependent G protein alphas guanine nucleotide exchange factor. J Biol Chem. 2011;286:19932–19942. doi: 10.1074/jbc.M110.163675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates JC. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 2003;13:463–471. doi: 10.1016/s0962-8924(03)00167-3. [DOI] [PubMed] [Google Scholar]
- Costa M, Wilson ET, Wieschaus E. A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell. 1994;76:1075–1089. doi: 10.1016/0092-8674(94)90384-0. [DOI] [PubMed] [Google Scholar]
- Couwenbergs C, Spilker AC, Gotta M. Control of embryonic spindle positioning and Galpha activity by C. elegans RIC-8. Curr Biol. 2004;14:1871–1876. doi: 10.1016/j.cub.2004.09.059. [DOI] [PubMed] [Google Scholar]
- David NB, Martin CA, Segalen M, Rosenfeld F, Schweisguth F, Bellaïche Y. Drosophila Ric-8 regulates Galphai cortical localization to promote Galphai-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat Cell Biol. 2005;7:1083–1090. doi: 10.1038/ncb1319. [DOI] [PubMed] [Google Scholar]
- Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, Wieschaus EF. Folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132:4165–4178. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- Dhingra A, Sulaiman P, Xu Y, Fina ME, Veh RW, Vardi N. Probing neurochemical structure and function of retinal ON bipolar cells with a transgenic mouse. J Comp Neurol. 2008;510:484–496. doi: 10.1002/cne.21807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech C, Patrikainen L, Kerr DS, Grall S, Liu Z, Laugerette F, Malnic B, Montmayeur JP. Ric-8A, a Galpha protein guanine nucleotide exchange factor potentiates taste receptor signaling. Front Cell Neurosci. 2009;3:11. doi: 10.3389/neuro.03.011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa M, Hinrichs MV, Bunster M, Babbitt P, Martinez-Oyanedel J, Olate J. Biophysical studies support a predicted superhelical structure with Armadillo repeats for Ric-8. Protein Sci. 2009;18:1139–1145. doi: 10.1002/pro.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay M, Pinter ME, Wright FA, Chan P, Murphy AJ, Valenzuela DM, Yancopoulos GD, Tall GG. Ric-8 proteins are molecular chaperones that direct nascent G protein α subunit membrane association. Sci Signal. 2011;4:ra79. doi: 10.1126/scisignal.2002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohla A, Offermanns S, Wilkie TM, Schultz G. Differential involvement of Galpha12 and Galpha13 in receptor-mediated stress fiber formation. J Biol Chem. 1999;274:17901–17907. doi: 10.1074/jbc.274.25.17901. [DOI] [PubMed] [Google Scholar]
- Grandy R, Sepulveda H, Aguilar R, Pihan P, Henriquez B, Olate J, Montecino M. The Ric-8B gene is highly expressed in proliferating preosteoblastic cells and downregulated during osteoblast differentiation in a SWI/SNF- and C/EBPbeta-mediated manner. Mol Cell Biol. 2011;31:2997–3008. doi: 10.1128/MCB.05096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampoelz B, Hoeller O, Bowman SK, Dunican D, Knoblich JA. Drosophila Ric-8 is essential for plasma-membrane localization of heterotrimeric G proteins. Nat Cell Biol. 2005;7:1099–1105. doi: 10.1038/ncb1318. [DOI] [PubMed] [Google Scholar]
- Hinrichs M, Torrejón M, Montecino M, Olate J. Ric-8: different cellular roles for a heterotrimeric G-protein GEF. J Cell Biochem. 2012;113:2797–2805. doi: 10.1002/jcb.24162. [DOI] [PubMed] [Google Scholar]
- Kanesaki T, Hirose S, Grosshans J, Fuse N. Heterotrimeric G protein signaling governs the cortical stability during apical constriction in Drosophila gastrulation. Mech Dev. 2013;130:132–142. doi: 10.1016/j.mod.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Kerr DS, Dannecker Von LEC, Davalos M, Michaloski JS, Malnic B. Ric-8B interacts with G alpha olf and G gamma 13 and co-localizes with G alpha olf, G beta 1 and G gamma 13 in the cilia of olfactory sensory neurons. Mol Cell Neurosci. 2008;38:341–348. doi: 10.1016/j.mcn.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Kölsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315:384–386. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol. 2007;8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- Leptin M. Drosophila gastrulation: from pattern formation to morphogenesis. Annu Rev Cell Dev Biol. 1995;11:189–212. doi: 10.1146/annurev.cb.11.110195.001201. [DOI] [PubMed] [Google Scholar]
- Li Y, Yan X, Wang H, Liang S, Ma WB, Fang M-Y, Talbot NJ, Wang ZY. MoRic8 Is a novel component of G-protein signaling during plant infection by the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact. 2010;23:317–331. doi: 10.1094/MPMI-23-3-0317. [DOI] [PubMed] [Google Scholar]
- Luo X, et al. Characterization of gene expression regulated by American ginseng and ginsenoside Rg3 in human colorectal cancer cells. Int J Oncol. 2008;32:975–983. [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Agurto R, Toro G, Fuentealba J, Arriagada C, Campos T, Albistur M, Henriquez JP, Olate J, Hinrichs MV, Torrejón M. Cloning and spatiotemporal expression of RIC-8 in Xenopus embryogenesis. Gene Expr Patterns. 2011;11:401–408. doi: 10.1016/j.gep.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF. Integration of contractile forces during tissue invagination. J Cell Biol. 2010;188:735–749. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457:495–499. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Emerson MD, McManus JR, Rand JB. RIC-8 (synembryn): a novel conserved protein that is required for G(q)alpha signaling in the C. elegans nervous system. Neuron. 2000;27:289–299. doi: 10.1016/s0896-6273(00)00037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Rand JB. A role for RIC-8 (Synembryn) and GOA-1 (G(o)alpha) in regulating a subset of centrosome movements during early embryogenesis in Caenorhabditis elegans. Genetics. 2000;156:1649–1660. doi: 10.1093/genetics/156.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morize P, Christiansen AE, Costa M, Parks S, Wieschaus E. Hyperactivation of the folded gastrulation pathway induces specific cell shape changes. Development. 1998;125:589–597. doi: 10.1242/dev.125.4.589. [DOI] [PubMed] [Google Scholar]
- Muggerud AA, Edgren H, Wolf M, Kleivi K, Dejeux E, Tost J, Sørlie T, Kallioniemi O. Data integration from two microarray platforms identifies bi-allelic genetic inactivation of RIC8A in a breast cancer cell line. BMC Med Genomics. 2009;2:26. doi: 10.1186/1755-8794-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidou KK, Barrett K. A Rho GTPase signaling pathway is used reiteratively in epithelial folding and potentially selects the outcome of Rho activation. Curr Biol. 2004;14:1822–1826. doi: 10.1016/j.cub.2004.09.080. [DOI] [PubMed] [Google Scholar]
- Nishimura A, Okamoto M, Sugawara Y, Mizuno N, Yamauchi J, Itoh H. Ric-8A potentiates Gq-mediated signal transduction by acting downstream of G protein-coupled receptor in intact cells. Genes Cells. 2006;11:487–498. doi: 10.1111/j.1365-2443.2006.00959.x. [DOI] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- Pistor S, Chakraborty T, Niebuhr K, Domann E, Wehland J. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 1994;13:758–763. doi: 10.1002/j.1460-2075.1994.tb06318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Häcker U, Turck C, Vale RD. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr Biol. 2004;14:1827–1833. doi: 10.1016/j.cub.2004.09.078. [DOI] [PubMed] [Google Scholar]
- Ross EM. Coordinating speed and amplitude in G-protein signaling. Curr Biol. 2008;18:R777–R783. doi: 10.1016/j.cub.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall GG, Gilman AG. Resistance to inhibitors of cholinesterase 8A catalyzes release of Galphai-GTP and nuclear mitotic apparatus protein (NuMA) from NuMA/LGN/Galphai-GDP complexes. Proc Natl Acad Sci USA. 2005;102:16584–16589. doi: 10.1073/pnas.0508306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall GG, Krumins AM, Gilman AG. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J Biol Chem. 2003;278:8356–8362. doi: 10.1074/jbc.M211862200. [DOI] [PubMed] [Google Scholar]
- Thomas CJ, Briknarová K, Hilmer JK, Movahed N, Bothner B, Sumida JP, Tall GG, Sprang SR. The nucleotide exchange factor Ric-8A is a chaperone for the conformationally dynamic nucleotide-free state of Gαi1. PLoS One. 2011;6:e23197. doi: 10.1371/journal.pone.0023197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CJ, Tall GG, Adhikari A, Sprang SR. Ric-8A catalyzes guanine nucleotide exchange on G alphai1 bound to the GPR/GoLoco exchange inhibitor AGS3. J Biol Chem. 2008;283:23150–23160. doi: 10.1074/jbc.M802422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tõnissoo T, Lulla S, Meier R, Saare M, Ruisu K, Pooga M, Karis A. Nucleotide exchange factor RIC-8 is indispensable in mammalian early development. Dev Dyn. 2010;239:3404–3415. doi: 10.1002/dvdy.22480. [DOI] [PubMed] [Google Scholar]
- Tõnissoo T, Meier R, Talts K, Plaas M, Karis A. Expression of ric-8 (synembryn) gene in the nervous system of developing and adult mouse. Gene Expr Patterns. 2003;3:591–594. doi: 10.1016/s1567-133x(03)00119-4. [DOI] [PubMed] [Google Scholar]
- Von Dannecker LEC, Mercadante AF, Malnic B. Ric-8B, an olfactory putative GTP exchange factor, amplifies signal transduction through the olfactory-specific G-protein Galphaolf. J Neurosci. 2005;25:3793–3800. doi: 10.1523/JNEUROSCI.4595-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Dannecker LEC, Mercadante AF, Malnic B. Ric-8B promotes functional expression of odorant receptors. Proc Natl Acad Sci USA. 2006;103:9310–9314. doi: 10.1073/pnas.0600697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Guo D, Xing B, Zhang JJ, Shu HB, Guo L, Huang XY. Resistance to inhibitors of cholinesterase-8A (Ric-8A) is critical for growth factor receptor-induced actin cytoskeletal reorganization. J Biol Chem. 2011;286:31055–31061. doi: 10.1074/jbc.M111.253427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ng KH, Qian H, Siderovski DP, Chia W, Yu F. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat Cell Biol. 2005;7:1091–1098. doi: 10.1038/ncb1317. [DOI] [PubMed] [Google Scholar]
- Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol Cell Biol. 2010;30:3519–3530. doi: 10.1128/MCB.00394-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SJ, Inchausti R, Eaton CJ, Krystofova S, Borkovich KA. RIC8 is a guanine-nucleotide exchange factor for Galpha subunits that regulates growth and development in Neurospora crassa. Genetics. 2011;189:165–176. doi: 10.1534/genetics.111.129270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Bradley L, Ambdukar I, Gutkind JS. A mutant alpha subunit of G12 potentiates the eicosanoid pathway and is highly oncogenic in NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:6741–6745. doi: 10.1073/pnas.90.14.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa SI. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J Biol Chem. 1998;273:32353–32359. doi: 10.1074/jbc.273.48.32353. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, Touhara K. Myr-Ric-8A enhances G(alpha15)-mediated Ca2+ response of vertebrate olfactory receptors. Chem Senses. 2009;34:15–23. doi: 10.1093/chemse/bjn047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.