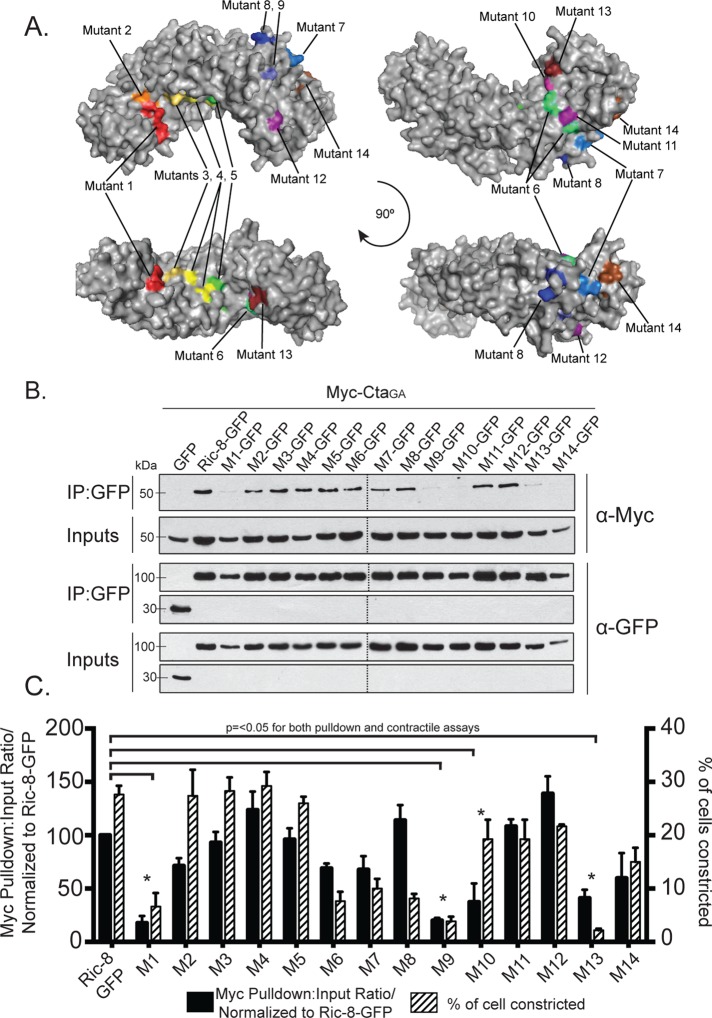
FIGURE 5:
Evolutionarily conserved electrostatic residues are required for binding between Ric-8 and CtaGA. (A) Clusters of point mutants used in our screen are represented by different colors on a model of Ric-8. (B, C) Mutant clusters within Ric-8 disrupt binding to CtaGA and inhibit pathway activation downstream of Fog. (B) S2 cells were transfected with GFP, Ric-8–GFP, or cluster Ric-8–GFP mutants and CtaGA. IPs were performed with GFP-binding protein and probed with anti-GFP and anti-Myc. (C) The pull-down:input ratios for Ric-8–GFP and Myc-CtaGA were quantified using densitometry and normalized to Ric-8–GFP (±SEM; error bars, p < 0.05; black bars). S2R+ cells were depleted of endogenous Ric-8 and transfected with Ric-8–GFP and cluster Ric-8–GFP mutants and then scored for the percentage of transfected cells constricting within the population (±SEM; error bars, p < 0.05; hatched bars). Dashed lines indicate where two separate gels have been combined.