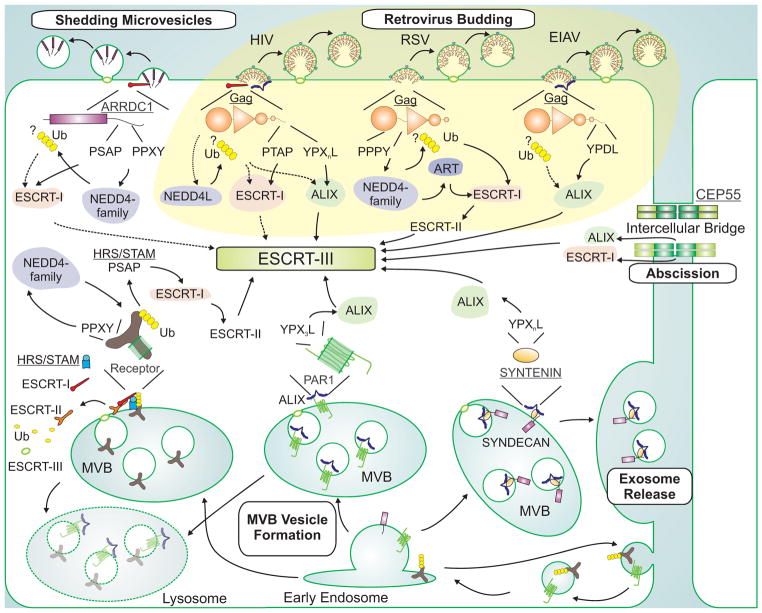
Figure 1. ESCRT pathway recruitment.
Cellular and viral adaptor proteins and complexes that recruit early-acting ESCRT factors and NEDD4 family ubiquitin E3 ligases to different sites of ESCRT-dependent membrane fission are depicted schematically. The figure emphasizes how different retroviruses hijack the ESCRT pathway using late assembly domains within their Gag polyproteins (yellow background, Gag proteins in orange, with their constituent domains shown schematically), and how these interactions mimic analogous interactions between cellular adaptors (underlined) and ESCRT factors (white background). Polyubiquitin chains are depicted as connected yellow hexagons, solid arrows denote known protein-protein interactions, dashed arrows denote inferred or indirect protein-protein interactions, and question marks indicate that the site(s) of ubiquitin attachment are uncertain (see text). Abbreviations: HIV, Human Immunodeficiency Virus; RSV, Rous sarcoma virus; EIAV, Equine Infectious Anemia Virus; MVB, multivesicular body; ART, arrestin-related trafficking adaptor. For illustration purposes, we selected retroviruses that bud primarily through P(T/S)AP (HIV-1), PPXY (RSV) and YPXL (EIAV) late assembly domains, but note that these viruses all also use auxiliary late assembly domains, including an YPXL motif in RSV (not shown) (Dilley et al., 2010).