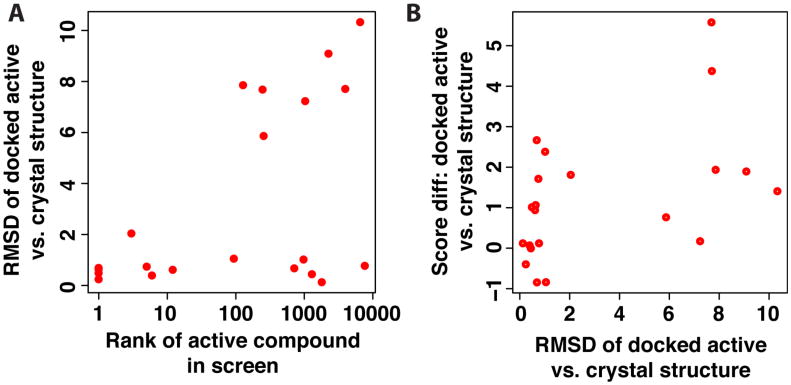
Figure 4.
(A) For each of the active compounds in the PPI set, the root mean square deviation (RMSD) of the docked active compound is computed relative to the crystal structure, and reported as a function of the rank of the active compound in the virtual screening experiment. Approximately half the compounds that were not ranked highly were mis-docked (high RMSD), while the other half were correctly docked but still did not rank well relative to the decoy compounds (low RMSD but high rank). (B) The difference in score between the docked active compound and the crystal structure is shown as a function of the RMSD. The mis-docked structures scored better than the crystal structure in all cases (positive score differences), suggesting that the energy function did not provide the correct relative ranking of these two poses.