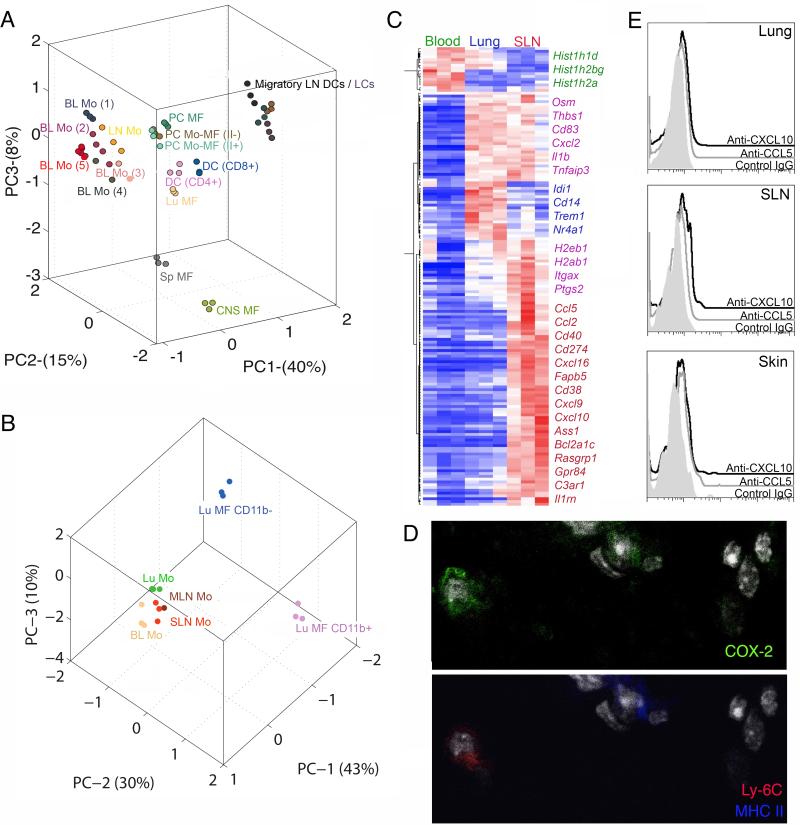
Figure 3. Gene expression analysis comparing tissue and blood monocytes with macrophages and DCs.
Principle component analysis (PCA) is a mathematical procedure that depicts uncorrelated variables on each axis. Percentage shown on each axis indicate the percent of variability explained, with the first priniciple component (PC1) defined as that which explains the most variability. The placement of data in proximity indicates a closer relationship between the datasets. A) PCA of LN resident DCs CD8+ and CD4+, migratory LN DCs, Langerhans cells (LC), macrophages (MF), and monocytes (Mo) from LN and blood (BL) (population gates 1-5 from Fig. 2; gate numbers next to Mo symbol). PC, peritoneal cavity; CNS, central nervous system; Sp, spleen; Lu, lung. Each population is depicted with a different color. B) PCA of lung (Lu) alveolar macrophages (CD11b−) and interstitial macrophages (CD11b+), and monocytes from blood, lung, MLN and SLN. Each population is depicted with a different color. C) Heat map depicts genes that are elevated ≥ 3-fold in blood, lung and SLN monocytes. D) Immunostaining for COX-2 expression in Ly6C+ monocytes and DCs of the lung parenchyma. E) Intracellular staining for chemokines, CXCL10 (open black) and CCL5 (open gray), along with isotype control (shaded gray) in the lung, skin and SLN. Fig. S3 and Tables S1 and S2 accompany this figure.