Figure 1.
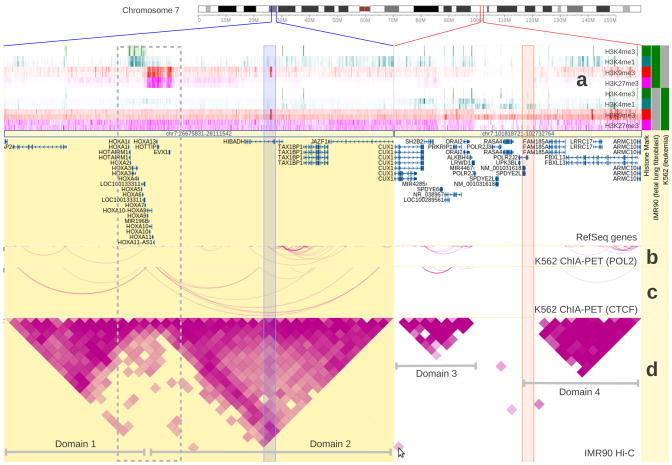
Two genomic regions (marked by blue or red lines on the chromosome ideogram) with stable chromatin domains are characterized by cell type-specific epigenetic profiles. The chromatin domains are marked by three long-range interaction tracks on two cell types. (a) Heatmap view of histone modification profiles of both IMR90 and K562 cells. (b) and (c) are two ChIA-PET tracks from K562 cells with different DNA binding proteins (POL2 and CTCF respectively, ENCODE data). (d) is a Hi-C track from IMR90 cells5. The triangle shapes in the Hi-C track depict chromatin domains in IMR90 cells (labeled Domains 1, 2, 3, 4), and the arcs in the ChIA-PET tracks indicate similar domain structure in K562 cells, suggesting that chromatins in these regions are organized similarly in both IMR90 and K562 cells. Histone modification tracks are shown in distinct colors to indicate the modification (red and fuchsia for repressive histone marks, green and teal for active histone marks). Histone modification marks are also identified by the color blocks in the metadata color map on the right. In the first region, the HOXA gene cluster at the boundary of the two domains shows a bipartite histone modification pattern (enclosed by a grey dotted box) in IMR90 cells. The left part of the cluster is covered by active marks (H3K4me3 and H3K4me1) and the right part by repressive marks (H3K9me3 and H3K27me3). This is consistent with the normal, co-linear expression pattern of this locus. In K562 cells this pattern is replaced by a uniform repressive pattern (depleted of active histone marks and enriched in repressive H3K27me3 mark). In the second region, IMR90 and K562 cells do not show significant differences in their histone modification patterns. A cell in the Hi-C track suggests an interaction event between the two regions (the spot is indicated by the cursor, the underlying interacting regions are indicated by the two semi-transparent columns). This cell represents a highly probable interaction event between two remote genomic regions. Additional information is provided in Supplementary Notes.