Figure 1.
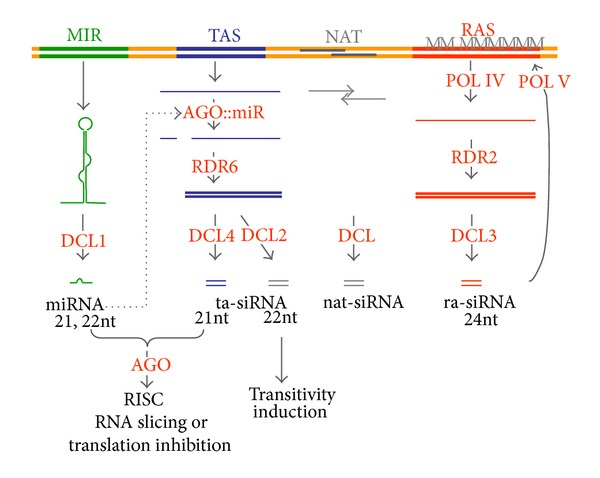
Posttranscriptional and transcriptional gene silencing pathways. Pri-miRNAs consist of a bulged hairpin flanked by unstructured arms. They are transcribed from the relevant MIR genes and are processed predominantly by the DCL1 “Drosha activity” and further by the DCL1 “Dicer activity,” yielding a miRNA duplex. Before processing, the pri-miRNAs, which can be extremely long, are spliced [46]. The DCL1 cofactor, the double-stranded RNA binding protein HYL1, and the 2′-OH ds RNA methyl transferases HEN1, SERRATE, or DAWDLE are not shown [34, 47–51]. Methylation by HEN1serves to protect the miRNA duplex from uridylation and degradation by SDN nucleases [52–57]. With miRNA guide strand and AGO1, a RISC is formed [58, 59], which binds to the cognate target and either slices it or arrests its translation. This step involves the function of cyclophilin 40 and HSP90 [60, 61]. TAS RNAs are transcribed from specific genes too, namely, the TAS genes. TAS RNAs are originally capped and polyadenylated, but they loose the cap and mostly also the poly-A end upon miRNA guided cleavage. They then become processed mainly by DCL4 in a phased way to generate secondary siRNAs, termed ta-siRNAs, which control target mRNAs, similarly as miRNAs [19, 62]. Nat-siRNAs are produced from overlapping dsRNA regions formed by natural antisense transcripts (NATs). Repeat-associated sequences (RASs) give rise to 24nt long ra-siRNAs through the action of DCL3. ra-siRNAs are amplified by POL IV and RDR2 and are involved in DNA and histone methylation by the action of POL V, AGO4, methylases, and chromoproteins [21].
