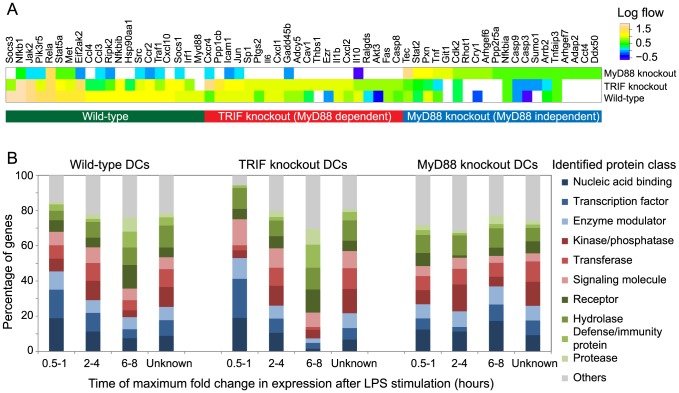
Figure 4. Comparison of MyD88 and TRIF-knockout gene sub-networks.
A. Comparison of the predicted flows of 20 genes from the optimal sub-networks of wild-type, MyD88-knockout and TRIF-knockout DCs. The heat map shows the similarity between the wild-type and the TRIF-knockout response to LPS stimulation through the large number of common genes predicted with high flows. B. Functional classes of genes based on the time of their highest fold change in expression and identified in the optimal sub-networks identified for wild-type, TRIF-knockout and MyD88 DCs on activation by LPS. “Unknown” denotes predicted genes that do not show significant change in expression on activation. Distinct gene groups are activated at different time points during the innate immune response. Protein class “Others” includes cytoskeletal proteins, cell adhesion molecules, calcium-binding proteins, ligases, transfer-carrier proteins, oxidoreductases, extracellular matrix proteins, transporters, chaperones, structural proteins, membrane traffic proteins, transmembrane receptor regulators, lyases, isomerases, cell junction proteins, surfactants and storage proteins.