Fig. 2.
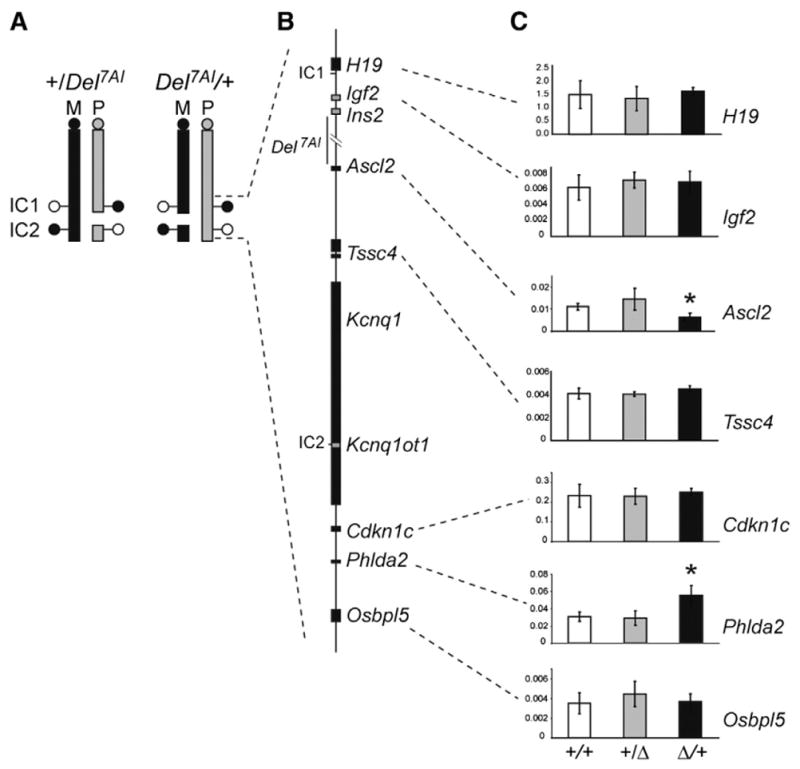
Total expression levels of IC1- and IC2-regulated imprinted genes in reciprocal heterozygous Del7AI placentae at E9.5. (A) Schematic representation of mouse chromosome 7 showing the breakpoints of the Del7AI allele relative to IC1 and IC2. DNA methylation status at the imprinting centers is shown by lollipops (white, unmethylated; black, methylated) on the maternal (M, black) and paternal (P, gray) homologues. (B) Map of the IC1 and IC2 clusters showing selective maternally (black) and paternally (gray) expressed genes. (C) qRT-PCR was performed on E9.5 placentae for the following genes: H19, Igf2, Ascl2, Tssc4, Cdkn1c, Phlda2 and Osbpl5. qRT-PCR was carried out as technical triplicates on three individual placentae (biological replicates) for each genotype and the error bars show the standard deviation for biological triplicates. Expression is relative to the reference housekeeping gene, Peptidylprolyl isomerase A (Ppia). Asterisks indicate mutant values significantly different from wild type values, using two-tailed unpaired Student t test (Ascl2, p = 0.006; Phlda2 p = 0.005).
