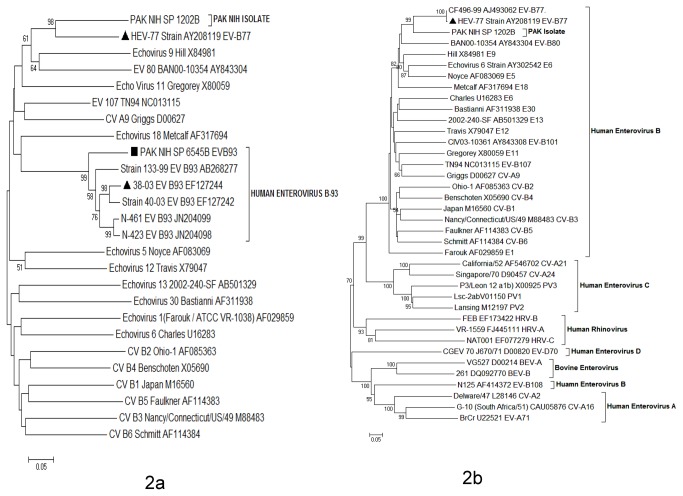
Figure 2. Unrooted Phylogenetic tree of Human Enteroviruses was constructed by Neighbor-joining (NJ) method with Kimura 2-parameter (K2-P) model using MEGA version 5.0 software.
This Phylogenetic tree based on A) VP1 and B) whole capsid (VP4-VP1) nucleotide sequences of PAK NIH isolates and other representative sequences of enterovirus serotypes retrieved from GenBank. AFP isolate is represented by ‘■’ taxon marker. The Prototype strain is labeled with taxon marker ‘▲’. Tree was evaluated with 1000 bootstrap pseudoreplicates. Bootstrap values greater than 50 are indicated at the respective nodes and the scale bar represents the evolutionary distance.