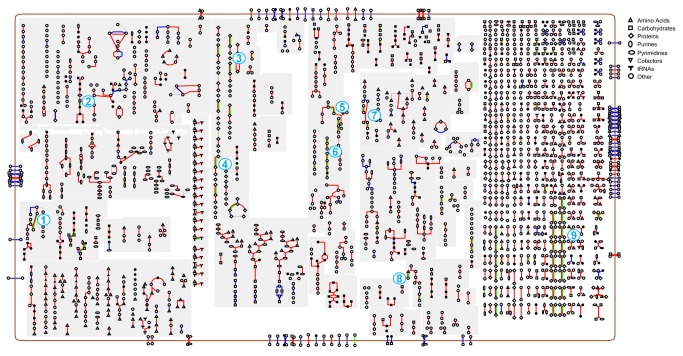
Figure 3. Overview of differentially expressed C. difficile pathways.
Pathways of C. difficile were reconstructed using pathway tools. Amino acids, carbohydrates, proteins, purines, pyrimidines, cofactors, tRNAs and other components in the pathways are coded as per the scheme given on the top right hand side of the figure. Pathways on the left side are biosynthetic pathways, middle are central intermediary metabolism pathways, right side are catabolic pathways and the group on the extreme right are reactions associated with the state of the cell. The following color scheme in the pathways represent; blue - present in at least one strain, red - present in all strains, green - differentially expressed in all strains. Numbers in the pathways denote the following differentially expressed pathways; 1- Gluconeogensis, 2 - Folate transformation, 3- Biotin-carboxyl carrier assembly, 4 - Plamitate biosynthesis, 5 - Pyruvate fermentation, 6 - Fermentation of butanote, 7- Lysine fermentation acetate, 8- Aminobutyrate degradation, 9- Degradation/Utilization/Assimilation.