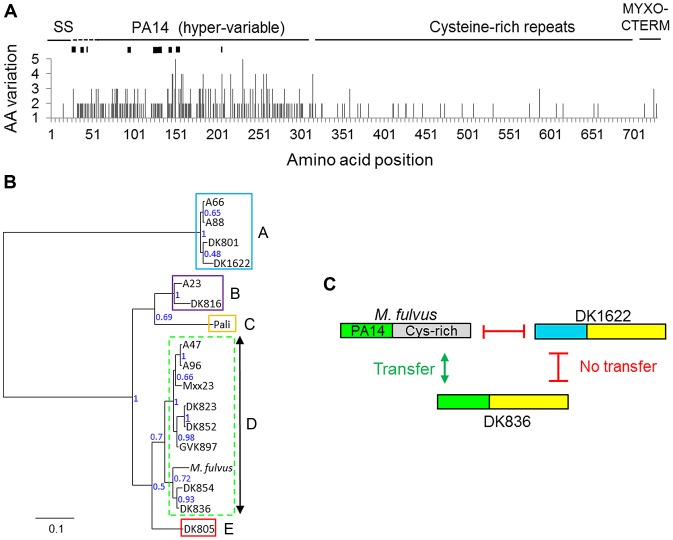
Figure 2. The TraA PA14-like domain is polymorphic and correlates to recognition groupings.
A) TraA amino acid (AA) variation derived from a sequence alignment from 16 M. xanthus strains is plotted. M. fulvus represents a distinct species and was excluded. Black rectangles in the hyper-variable region represent indels that range from one to seven codons in length. The signal sequence (SS) and a putative protein sorting tag (MYXO-CTERM) are also labeled. B) Phylogenetic tree derived from the PA14 polymorphic region, unrooted. Node support values are given as posterior probabilities. The multiple-sequence alignment used to generate the tree is provided in Figure S1. Recognition groups are boxed and labeled. A dashed border indicates the heterogeneous recognition group. C) Domain similarity between three TraA sequences is graphically depicted and color coded. Gray and blue regions contain divergent sequences. Transfer compatibility of TraA variants is shown by green arrows (transfer) or red bars (no transfer). Specificity was determined by PA14 domain relatedness. The apparent chimeric domain architecture, depicting sequence relatedness, suggests that DNA rearrangements occurred between ancestral traA alleles. See Figure S2 for alignments.