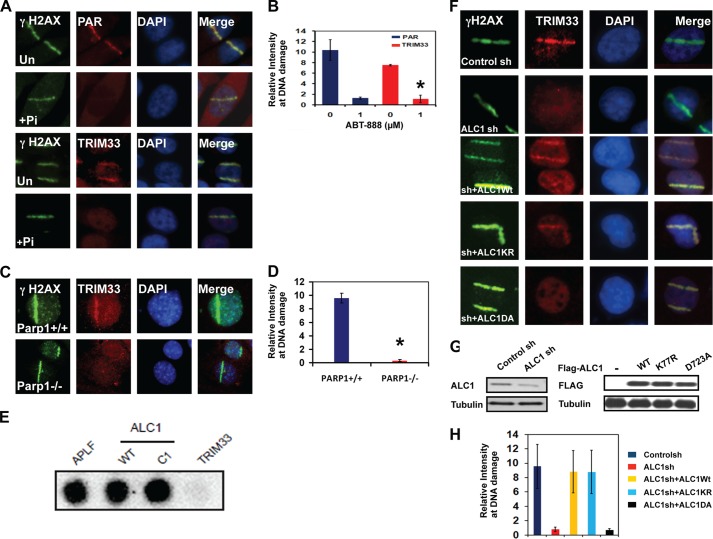
FIGURE 4.
Localization of TRIM33 to DNA breaks is dependent upon PARP and ALC1. A, PAR (top two panels) and TRIM33 (bottom two panels) were localized by IF in untreated (Un) and in cells pretreated with 1 μm PARPi (Pi) ABT-888 for 1 h. B, quantitation of PAR and TRIM33 colocalization with γH2AX at sites of laser scissors. *, p < 0.005. C, Parp1+/+ or Parp1−/− mouse embryonic fibroblasts were treated with laser scissors, and γH2AX and TRIM33 were localized by IF. Images are shown at identical magnification. D, quantitation of PAR and TRIM33 colocalization with γH2AX at sites of laser scissors. *, p < 0.005. E, APLF, WtALC1, C1 (ALC1 macro domain only), and TRIM33 proteins were dot-blotted onto a nitrocellulose membrane and incubated with 32P-labeled PAR. F, TRIM33 localization to DNA breaks is ALC1-dependent. U2OS cells stably expressing control sh, ALC1sh, or cells expressing ALC1 sh were reconstituted with WT ALC1, KR (kinase dead) and DA (PAR binding mutant) were analyzed. All cells were subjected to UV laser scissor-induced DNA breaks. After 10 min, cell were fixed, and IF was performed using antibodies to γH2AX and TRIM33. G, Western blot analyses showing levels of ALC1 and TRIM33 in U2OS cells expressing control and ALC1 shRNA and different constructs of ALC1 in ALC1sh cells. H, quantitation of relative intensity of TRIM33 at sites of DNA damage. Each data point is mean ± S.D. of at least 20 cells. *, p < 0.005.