FIGURE 2.
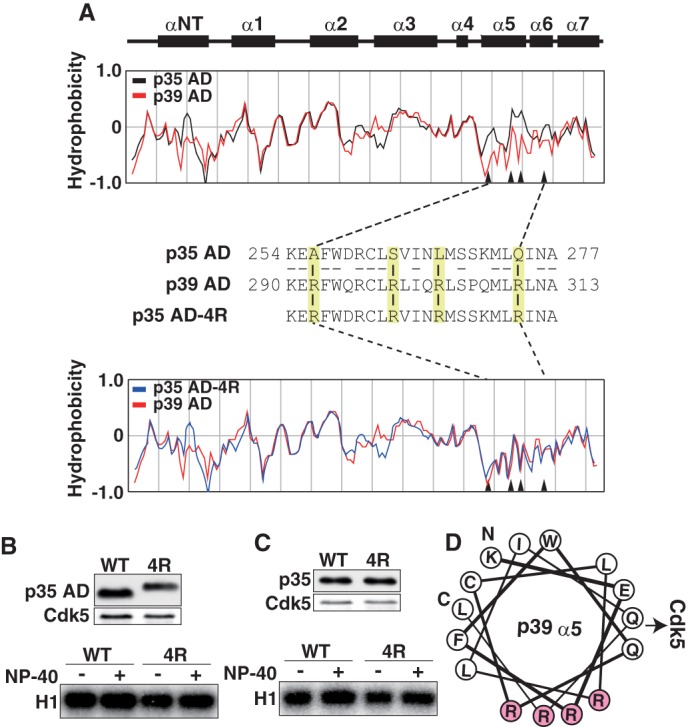
Different hydrophobicity of p35AD and p39 AD is not the cause of the different stability of Cdk5 complexes. A, the hydrophobicity of p35 AD and p39 AD. Upper two panels show the α-helices in p35 AD (23). The hydrophobicity of p35 AD (black) and p39 AD (red) is shown in the second panel. The third panel shows amino acid sequences of the α5-α6 region of p35 AD and p39 AD. Four amino acids in p35 AD, Ala-256, Ser-262, Leu-267, and Gln-274, are changed to arginine in p39 AD (yellow). The amino acid sequence of mutant p35 AD (p35 AD-4R) containing Arg at the above four sites is shown in the third line. The hydrophobicity of p35 AD-4R (blue) is compared with that of p39 AD (red) in the bottom panel. Arrowheads in hydrophobicity graphs indicate the four amino acids. B, kinase activity of p35 AD and p35 AD-4R. Expression of p35 AD (WT) or p35 AD-4R (4R) and Cdk5 in HEK293 cells was examined by immunoblotting with anti-FLAG (p35 AD) and anti-Cdk5 (Cdk5). p35 AD or p35 AD-4R was immunoprecipitated with anti-FLAG antibody. After washing in the absence (−) or presence (+) of 1% Nonidet P-40, the kinase activity of the immunoprecipitates was measured using histone H1 (H1) and [γ-32P]ATP as substrates (lower panel). C, kinase activity of p35 or p35–4R. Expression of p35 (WT) or p35–4R (4R) and Cdk5 in HEK293 cells is shown by immunoblotting with anti-FLAG (p35) or anti-Cdk5 (Cdk5). p35 or p35–4R was immunoprecipitated, and after washing in the absence (−) or presence (+) of 1% Nonidet P-40, the kinase activity was measured using histone H1 and [γ-32P]ATP as substrates (lower panel). D, diagram of the arrangement of amino acid residues in α5-helix, viewed down from the N-terminal Lys-290 (K) to the C-terminal Leu-311 (L) of the helix. All four Arg residues (R, red) are positioned in one side of the α-helix, perpendicular to the direction of Cdk5 binding.
