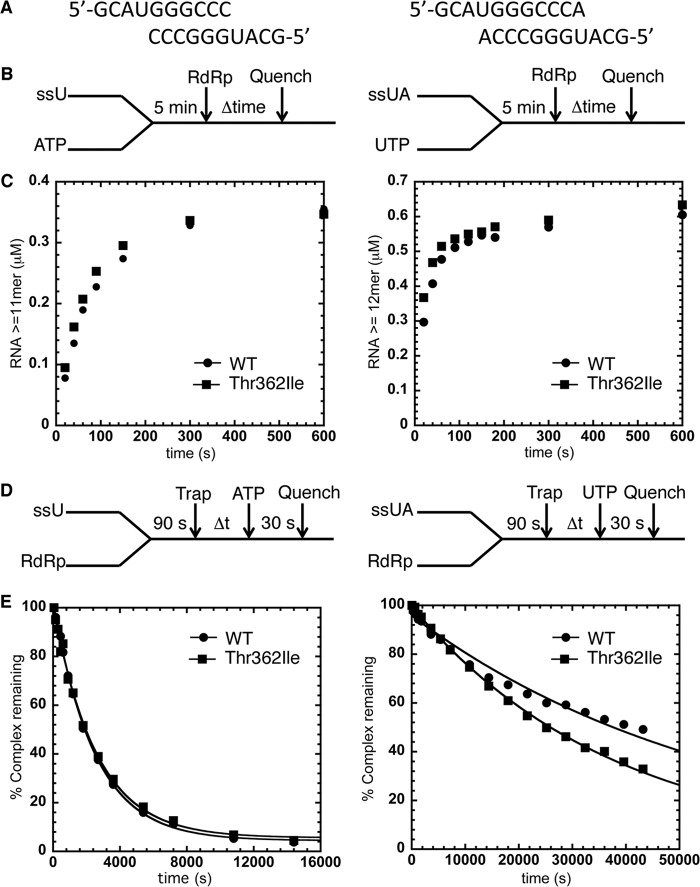
FIGURE 2.
Formation and stability of RdRp-RNA complexes is not affected by the T362I substitution in motif D. A, RNA sequences for sym/subU (left) and sym/subUA (right). B, experimental design for RdRp-RNA-NTP assembly assay. Reactions were initiated by adding RdRp (1 μm) into sym/subU or sym/subUA (0.5 μm duplex) and its corresponding ATP or UTP (500 μm), which were incubated at 30 °C for 5 min. At the indicated times, reactions were quenched by adding 25 mm EDTA. C, the RdRp-RNA-NTP assembly assay for WT RdRp (●) and T362I RdRp (■) using both sym/subU and sym/subUA. There is essentially no difference in this assay between WT and T362I RdRp. D, experimental design for the RdRp-RNA dissociation assay. RdRp and RNA were incubated at 30 °C for 90 s at which time trap (100 μm unlabeled RNA) was added to the reaction buffer. After the indicated times, the reaction buffer was mixed with ATP or UTP (500 μm) and then quenched after 30 s by adding EDTA (25 mm). E, the RdRp-RNA dissociation assay for WT RdRp (●) and T362I RdRp (■) using both sym/subU and sym/subUA RNA. The solid line is the data fit to a single exponential function. The kdiss(obs) for sym/subU (sym/subUA) was 4.00 ± 0.09 × 10−4 s−1 (1.70 ± 0.07 × 10−5 s−1) for WT RdRp and 3.90 ± 0.02 × 10−4 s−1 (2.70 ± 0.03 × 10−5 s−1) for T362I RdRp, indicating that the RNA binding characteristics of WT and T362I RdRp are nearly identical. Each data point is from the averages of at least two separate experiments on different days.