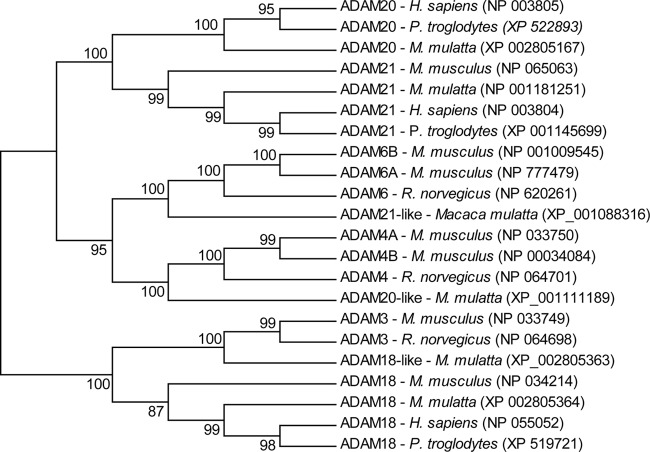
Fig. 3.
Maximum-likelihood phylogenetic analysis of Macaque ADAM-like proteins. Evolutionary history was inferred by using the Maximum Parsimony method. The bootstrap consensus tree (shown) was inferred from 1000 replicates (24). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The analysis involved 22 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 939 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (23).