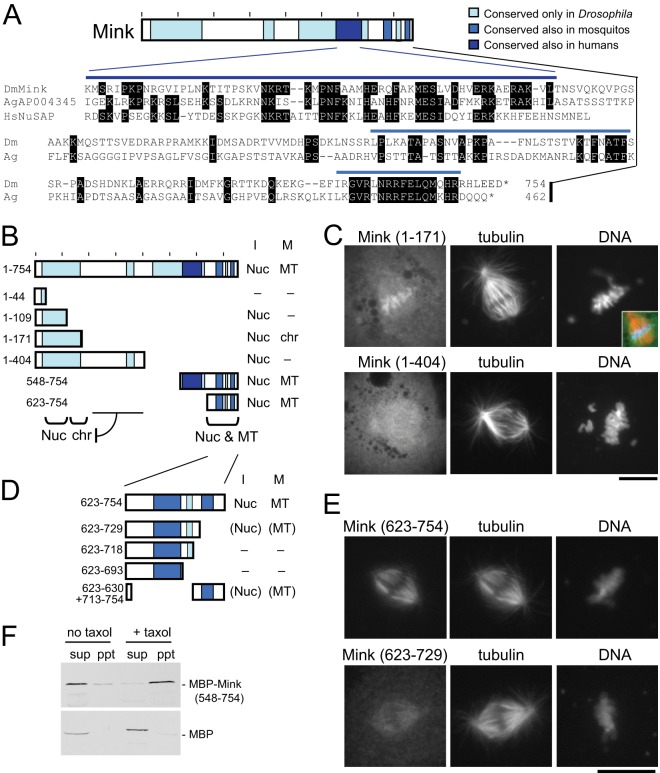
Fig. 4.
The domain structure of Mink. A, conserved regions of Mink and a protein sequence alignment among Mink, human NuSAP, and a mosquito protein (Anopheles gambiae AGAP004345). Identical residues are marked. B, a summary of the subcellular localization of truncated Mink proteins. “I” and “M” indicate interphase and mitosis. “Nuc,” “chr,” and “MT” indicate localization to nucleus, chromosomes, and microtubules, respectively. “–” indicates diffuse localization. C, immunostaining of mitotic S2 cells transiently expressing GFP-fused truncated Mink proteins using antibodies against GFP and α-tubulin. The inset shows a merged image with Mink in green, tubulin in red, and DNA in blue. Bar = 10 μm. D, a summary of the subcellular localization of truncated Mink proteins. Localization locations in parentheses indicate weak localization. E, immunostaining of mitotic S2 cells transiently expressing GFP-fused truncated Mink proteins using antibodies against GFP and α-tubulin. Bar = 10 μm. F, the C-terminal region of Mink directly interacts with microtubules in vitro. MBP-fused Mink (548–754) or MBP alone was produced in bacteria and incubated with tubulin. After microtubules had been polymerized using taxol and GTP, microtubules were spun down. For the control, taxol and GTP were omitted. Supernatants (sup) and pellets (ppt) were analyzed via Western blot using an antibody against MBP.