Abstract
We present GenoSuite, an integrated proteogenomic pipeline to validate, refine and discover protein coding genes using high-throughput mass spectrometry (MS) data from prokaryotes. To demonstrate the effectiveness of GenoSuite, we analyzed proteomics data of Bradyrhizobium japonicum (USDA110), a model organism to study agriculturally important rhizobium-legume symbiosis. Our analysis confirmed 31% of known genes, refined 49 gene models for their translation initiation site (TIS) and discovered 59 novel protein coding genes. Notably, a novel protein which redefined the boundary of a crucial cytochrome P450 system related operon was discovered, known to be highly expressed in the anaerobic symbiotic bacteroids. A focused analysis on N-terminally acetylated peptides indicated downstream TIS for gene blr0594. Finally, ortho-proteogenomic analysis revealed three novel genes in recently sequenced B. japonicum USDA6T genome. The discovery of large number of missing genes and correction of gene models have expanded the proteomic landscape of B. japonicum and presents an unparalleled utility of proteogenomic analyses and versatility of GenoSuite for annotating prokaryotic genomes including pathogens.
Rapid advances in massively parallel sequencing technologies have enabled the sequencing of thousands of prokaryote genomes. However, to understand the functional elements of the genome, annotation of the protein coding genes is the first and usually the most challenging task. Protein coding genes in a genome are annotated by in silico predictions and homology searches against known proteins. In silico gene annotations are prone to errors such as missing genes, incorrect translation initiation sites (TIS)1, and pseudogenes. For instance, TIS mis-annotations have been estimated to be around 10–40% in several bacterial and archaeal genomes (1, 2). Some genes are wrongly annotated as pseudogenes in bacterial genomes (3). Several proteogenomic studies have reported new protein coding genes which were previously missed by in silico gene annotations (4–6).
Mass spectrometry has emerged as a sensitive high-throughput method in which thousands of proteins can be identified in a single experiment (7). Peptides identified from mass spectrometry based proteomics data can be used as experimental evidence to identify protein coding genes and correct such annotation errors (7). The method of harnessing mass spectrometry proteomic data to annotate genomes is generally referred as proteogenomics (8). This approach has been successfully applied to re-annotate several genomes (1, 9–12) and to improve annotations of larger taxonomic groups than a single bacterium (1, 13). Additionally, identification of peptides with protein N-terminal modifications like formylated methionine or N-acetylation has been used to annotate TIS for genes (5). Despite its direct benefits, proteogenomic analyses is computationally challenging, multistep process and tends to have high rates of false positive identification (14). Multi-algorithmic search approaches have been shown to increase sensitivity and specificity in large scale proteomic studies (15, 16) but are difficult to carry out in a proteogenomic context. This is because of the lack of automated software for proteogenomic analyses that incorporates multiple search engines without compromising on the statistical robustness of individual algorithms.
To fill this gap, we developed an automated pipeline, GenoSuite to carry out genome translations, database searches using multiple search engines, result integration based on statistical significance of PSMs, FDR calculations, coordinate mapping, and finding completely novel genes. To demonstrate the effectiveness of GenoSuite, we analyzed proteomics data of Bradyrhizobium japonicum (USDA110), a model organism to study agriculturally important rhizobium–legume symbiosis. We selected B. japonicum for proteogenomic re-annotation because of its large genome size, high GC content and nonavailability of many closely related genome sequences. B. japonicum is a symbiont to legumes and is important for nitrogen fixation in root nodules which helps these plants to grow without any nitrogenous fertilizers. Its primary host is Soybean, an economically important crop and model system to study rhizobia-legume symbiosis. This bacterium has a 9.1 Mb genome, one of the longest among bacteria, with 64.1% GC content (17). Kaneko et al. annotated 8,317 protein coding regions by in silico approach based on Glimmer (18) gene predictions and sequence similarity with known proteins. A large number of transcriptomics and proteomics studies have also been performed to understand the mechanisms and genes involved in symbiosis and nitrogen fixing process. However, a high quality annotation of protein coding genes is still not achieved for this class of bacteria. To improve on the existing annotation of B. japonicum, we carried out a comprehensive proteogenomic analysis using publicly available proteomics data generated from bacteroids of three host systems (19, 20). We used GenoSuite to search spectral data against genome translated database of B. japonicum and selected peptide identifications at ≤1%FDR to re-annotate B. japonicum genome. We identified 59 novel protein coding regions (NPCRs) and corrected annotations for 49 genes.
EXPERIMENTAL PROCEDURES
Data
The mass spectral data for proteogenomic analysis on B. japonicum were obtained from PRIDE repository (21). A total of nine data sets with PRIDE accessions 10099–10104 and 10114–10116 were used. These mass spectra represent B. japonicum proteomes from three different host systems in triplicates for each host. In total, these data sets had 621,176 MS/MS spectra.
Development of GenoSuite, An Automated Proteogenomic Pipeline
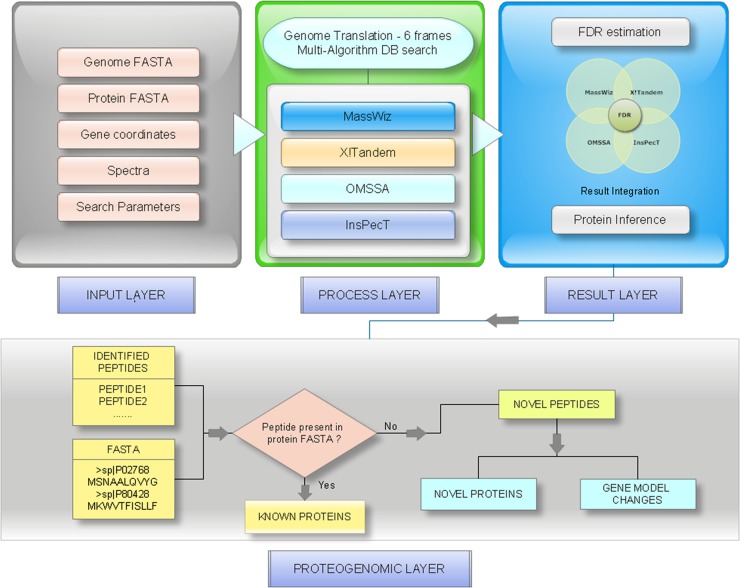
We developed GenoSuite, a standalone pipeline for automated proteogenomic analysis. GenoSuite is a suite of three tools: PPT (Prokaryotic Proteogenomic Tool), ORFmapper, and PSMplotter. GenoSuite is an easily configurable tool and is ready for use after downloading and unzipping the archive. Integration with the search algorithms OMSSA, X!Tandem, and InsPecT is also easy because only the paths need to be added to GenoSuite configuration file. MassWiz comes integrated as a part of the standard distribution.
PPT searches spectral data against a genome database with four peptide identification algorithms namely MassWiz (22), OMSSA (23), X!Tandem (24) and InsPecT (25). It is highly configurable and any combination of these algorithms can be used for search. All inputs for PPT are defined in a configuration file. GenoSuite uses common file formats e.g. FASTA, Genbank, MGF, GFF etc., so that the pipeline can be easily integrated with existing frameworks. We employed a Combined FDRScore (26) based approach to integrate results from multiple algorithms, each of which has a noncomparable scoring metric. In brief, GenoSuite calculates FDRScore for all employed algorithms' results. Average FDRScore is calculated for each PSM by calculating geometric mean of FDRScores from individual algorithms identifying peptide spectrum pair. All PSMs are divided into subsets based on the combination of algorithms identifying PSMs. Combined FDRScores are again calculated from Average FDRScores separately for each subset of PSMs and significant PSMs below a user defined FDR threshold are selected for further analyses. False Discovery Rate (FDR) (in %) is estimated by Kall method (27)
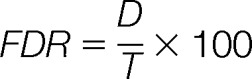 |
D = Decoy PSMs passing the threshold
T = Target PSMs passing the threshold
Filtered peptides are then mapped onto the genome and also onto the known proteins. Identified peptides mapping exclusively to the genome translated database are classified as novel peptides. A complete outline of the analysis pipeline GenoSuite is shown in Fig. 1. List of novel peptides are exported as GFF files which can be integrated with any DAS annotation server. For spectral quality assessment, XML files are created for peptide identifications from each algorithm and for novel peptides.
Fig. 1.
Schematic representation of GenoSuite workflow.
ORFmapper compares novel peptides to existing annotations and to ab initio predictions. It requires genbank file, ORF prediction file (GeneMark or GFF format) and novel peptide GFF as inputs. Novel peptide coordinates in input GFF file are compared with the gene coordinates in genbank file and peptides are further classified into (1) novel proteins coding region (NPCR) or (2) gene model changes. ORFmapper creates separate files for peptides leading to novel proteins, peptides suggesting gene model changes and ORFs mapped to novel peptides. It also creates genomic map for each peptide to provide a visual of its genomic context. These images are linked to an HTML file for ease of analysis.
PSMplotter program is a utility to visualize peptide spectrum matches. It takes the XML file created by PPT as its Input and generates an HTML file where all spectrum matches from the XML file are hyperlinked with their PSM images. This can be used for manual validation of peptide spectral matches.
It is written in Perl and the executables are distributed for Windows and Linux platforms. It is freely available for download at (https://sourceforge.net/projects/proteogenomic/files/). The code is open-source and freely available for academic purpose on request.
Proteogenomic Analysis
Spectral data were searched by GenoSuite against a six frame translated database of B. japonicum USDA110 genome (NC_004463.1). GenoSuite translates a genome from stop to stop codon in all six reading frames and translation products of length 50 aa or more are kept in database. In total, the B. japonicum database had 98,716 sequence entries. All four algorithms in the GenoSuite were employed for spectral data search with 20 ppm precursor ion tolerance, 0.6 Da product ion tolerance, trypsin as the protease with one missed cleavage, carbamidomethylation of cysteine as a fixed modification and methionine oxidation as variable modification. For parameters not common across algorithms, we used the algorithm defaults. Separate target-decoy database searches were carried out. Stringent FDR threshold of ≤1% was applied to the resulting PSMs. Leucine and Isoleucine were considered identical during FDR calculations and mapping onto the genome or proteome. Peptides mapping to two different genomic loci were not considered for further analysis.
Translation Initiation Site Search
A different search for estimating translation initiation sites for the gene models was performed using OMSSA and X!Tandem. Search parameters were same as above except semitryptic enzyme specificity, peptide N-terminal acetylation and peptide N-terminal formylated methionine as variable modifications. Only N-terminally modified peptides identified at ≤ 1% FDR were selected for further analysis.
Gene Prediction and ORF Comparison
To define boundaries of maximum number of novel translations in the B. japonicum USDA110 genome we used ORF predictions by four different algorithms namely GeneMark.hmm prokaryotic (Version 2.6r) (28), Glimmer3 (18), Prodigal.v2.50 (29), and FGENESB (30). GeneMark, Glimmer, and Prodigal predictions were downloaded from NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Bradyrhizobium_japonicum_USDA_110_uid57599/). FGENESB gene predictions were downloaded from (http://linux1.softberry.com/data/annotation/bact/NC_004463.fna.ann.gz). ORF comparison across algorithms was performed using in-house scripts.
Protein Functional Annotation
To annotate proteins for their probable functions, we used domain assignment method Pfam (31), Gene ontology assignments based on sequence similarity by GOANNA (32) and annotations by RAST (33). Pfam-A profile database and HMMER-3.0 were used to assign profiles. GOANNA tool was used for gene ontology assignment and only molecular function (F) category was used for annotation.
RESULTS
Multi-algorithmic Proteogenomic Analysis Pipeline Development
GenoSuite was developed to automate spectral searches, statistical integration of PSMs and downstream analysis. It is configured with four open source peptide identification algorithms namely OMSSA, X!Tandem, InsPecT and MassWiz. The choice of algorithms was empirical and these algorithms have been benchmarked in our previous studies (16, 22). These algorithms differ in their basic scoring and have their own advantages. For instance, OMSSA uses a Poisson distribution for separating significant matches from random hits (23) whereas X!Tandem uses a hyper-geometric model (24). MassWiz is an empirical scorer based on continuity, mass error, and intensity of matched fragment ions (22). InsPecT is a tag based peptide identification method (25). Although there are rescoring algorithms to improve database search results (34–36), a diverse set of algorithms provides an increased sensitivity for peptide identifications than any single algorithm. GenoSuite incorporates a reversed protein decoy database strategy to calculate FDR. For result integration from these algorithms, GenoSuite calculates Combined FDRScore which is reported to give ≈35% more identifications than any single algorithm (26). To check the consistency of Combined FDRScore calculation implemented in GenoSuite, we compared the combined FDRScores calculated by GenoSuite and FDRapp (37) from OMSSA and X!Tandem search results. We get highly correlated CFS values from these two tools (supplemental Fig. S1). FDRapp implements the same approach on OMSSA, X!Tandem, and Mascot algorithms. We extended this approach to Inspect and MassWiz and implemented in GenoSuite (supplemental Fig. S2). Peptides from FDR filtered PSMs are then mapped onto the genome to report NPCR and gene model changes. Fig. 1 depicts a schematic chart of GenoSuite. Additional functionality for ORF prediction mappings and peptide spectral match visualization is also added to the pipeline.
Genome Search Specific Peptide Identification
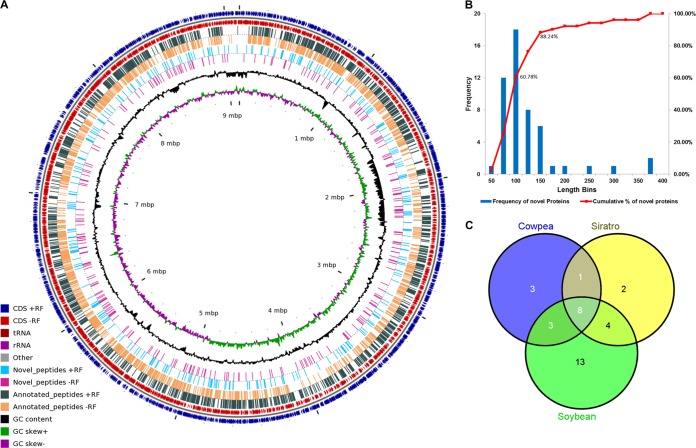
A total of 621,176 spectra from nine different datasets were searched against B. japonicum genome. A complete list of significant PSM identifications from each of nine datasets is provided in supplementary File S1. Total 26,592 peptides were identified of which 511 were shared across multiple genomic loci. As the primary aim of this study was to annotate genomic loci, shared peptides were not considered for further analysis. Fig. 2A depicts all unique peptides (26,081) identified in this study mapped to their genomic loci. After mapping unique peptides onto the genome translated database, GenoSuite reports proteins identified by at least two unique peptides or single peptide with ≥5 significant PSMs above desired FDR threshold. A total of 2654 proteins were identified in an automated manner at a global protein level FDR of 0.0018. Supplementary File S2 provides a full list of proteins identified in this analysis and their sample wise identification. Four out of ten proteins, which were identified with only a single unique peptide, had dubious peptide spectral matches and hence were not considered for further analysis. Supplementary File S3 provides annotated peptide spectral matches for six accepted single peptide hits. GenoSuite further maps all peptides from significant protein identifications to the annotated proteome and lists novel peptides. A total of 283 peptides were reported as novel. GenoSuite then categorizes novel peptides into (1) NPCR or (2) changes in the annotated gene models. A total of 59 new protein coding regions and 49 wrongly annotated genes were reported in this automated analysis by GenoSuite. A potential contamination of host proteins or any other contamination may also result in observation of novel peptides. To eliminate this possibility, we mapped all the novel peptides on a sequence database of Glycine max and common contaminants (cRAP) allowing one amino acid mismatch. Eighteen of these novel peptides, all of length 8 aa or below, were found to be similar with host or contaminant proteins. This did not affect the results at protein level as novel proteins had other unique peptides. One gene model change suggested exclusively by a peptide having similarity with host or contaminant proteins was not considered further.
Fig. 2.
Proteogenomic identifications of peptides and proteins. A, Genomic view of peptides identified. B, Length distribution of novel proteins. C, Host specific expression of novel proteins of B. japonicum.
Novel Protein Coding Regions in B. japonicum Genome
We discovered 59 confident NPCRs in B. japonicum USDA110 genome. Supplementary File S4 lists all NPCRs identified in this study with information about their function, sample-wise and host-wise expression. Ab initio gene predictions were used to determine the boundary of un-annotated genes. ORF predictions provide information about conservation of transcription and translational features beyond coding region and add confidence to the peptide identifications. Fifty-one out of 59 NPCRs are supported by ORF predictions on the same frame and strand. Based on gene predictions, we determined the TIS of these proteins. We also analyzed the length distribution of these 51 NPCRs. The average length of these proteins is 113 aa. Interestingly, 92% of these 51 novel proteins were below 200 aa length suggesting that most novel proteins are encoded by short ORFs (Fig. 2B). Supplemental Table S1 lists all NPCRs with their genomic coordinates and identified peptides. As our proteogenomic analysis is based on the proteomic data from different hosts of B. japonicum, we analyzed the NPCRs for their host-specific expression. Following the criterion of Koch et al. for this analysis, we considered only those NPCRs which are identified in at least two replicates for a host. Collectively, 34 NPCRs are identified in at least two replicates. 13 of these are specifically expressed in bacteroids from soybean, two from siratro and three are specific for cowpea bacteroids (Fig. 2C). Host specific expression of NPCRs suggests their putative role in host adaptation and survival.
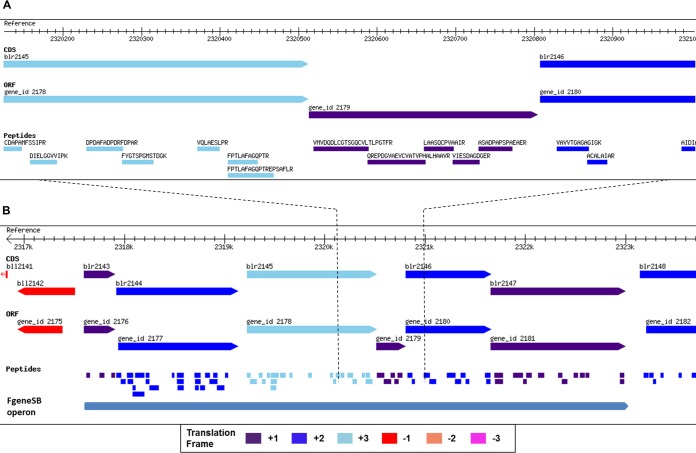
We also probed the genomic context of the NPCRs. Among all NPCRs, 36 were intergenic, 21 were on the opposite strand to existing gene annotation and two were on different frame. Fig. 3A shows a NPCR identified with five peptides mapping to an intergenic region of the genome. The protein is supported by ORF prediction by all four gene prediction algorithms and is identified in five out of nine samples. Function prediction suggests that this protein is a putative cytochrome P450 hydroxylase. Interestingly, Inclusion of this NPCR suggests a continuous operon in this genomic region (Fig. 3B). Earlier this operon was considered with only three members (blr2143-blr2145) but now with NPCR bridging the gap, this operon can be extended to six members. FGENESB also predicts an operon of six proteins, which includes the NPCR identified in this genomic locus. Sequence similarity shows the NPCR and operon to be specific to the rhizobia. The operon has been shown to be under regulation of oxygen-responsive transcriptional activator protein NifA (38) and is highly expressed in anaerobic conditions of bacteroids within the host (39). Four proteins of the operon are annotated as cytochrome P450 proteins and remaining two have cytochrome P450 hydroxylase domain. In our analysis, all six proteins of this operon are identified with multiple peptide hits. Identification of all proteins of this operon and their related functions further strengthens coordinated gene expression as in operonic structures.
Fig. 3.
Novel protein coding gene discovered in its genomic context. Reference track shows genomic co-ordinates. CDS track represent annotated protein coding genes. ORF track shows ORF predictions by GeneMark algorithm. Peptide track shows all identified peptides with their respective sequences. Different bar colors represent different reading frames. A, Five peptides map to a genomic region where no protein coding gene is annotated. GeneMark predicts a putative gene in the same translation frame of the identified peptide. B, The identified novel protein suggests a continuous operon. FGENESB also predicts an operon of six proteins including the novel protein. Peptides are identified for all proteins in the operon.
Novel Proteins and Comparative Genomics
We extended newly identified annotations of B. japonicum in related rhizobial genomes based on orthology of NPCRs. Ortho-proteogenomic strategy was first applied on genus Mycobacterium by Gallien et al. (1) and recently adopted by Christie-Oleza et al. for annotating Roseobacter clad (10). For this analysis we included all the sequenced genomes in Bradyrhizobiaceae family along with Mesorhizobium loti, Sinorhizobium meliloti, and Rhizobium leguminosarum because these are widely studied rhizobial genomes. These genomes are of diverse sizes ranging from ≈3 Mb to ≈9 Mb with 3122 to 8826 protein coding genes. Genomic level identity among these genomes and B. japonicum USDA110 genome ranges from 1% to 75% (supplemental Table S2). A full list of NPCRs with presence/absence of orthologs in related genomes is tabulated in supplementary File S5. Orthologous sequences were found for 43 NPCRs. Interestingly, orthologs of 35 NPCRs identified in this study in B. japonicum USDA110 are annotated as protein coding genes in the recently sequenced and annotated B. japonicum USDA6T (40) and/or B. sp.S23321 (41) genomes. After comparing the orthologous loci with genome annotations and ORF predictions of the respective genomes, we added three novel proteins in B. japonicum USDA6T genome and found three protein coding genes with N-terminal changes.
N-terminal Changes in the Annotated Genes
Sixty two novel peptides mapped upstream of annotated gene models on the same frame and strand. Because there was no stop codon between identified peptide and annotated gene, these peptides suggested change in the currently annotated translation initiation site (TIS). We excluded one peptide from the list because of its similarity with host proteins. The remaining 61 novel peptides indicated changes in 48 annotated gene models. A detailed list of peptides suggesting gene model changes is provided as supplementary File S6. The ORF predictions were checked for any upstream TIS. 34 of these 48 gene model changes are also supported by gene predictions. We could assign new TIS to 34 genes, 21 of which had TTG as the start codon. We considered maximum voted gene start of the ORF predictions and the longest ORF in cases where votes tied. Fig. 4 highlights a case of gene model change. Four peptides map upstream to the currently annotated gene model for locus bll2380, a gene coding for a glycosyl transferase enzyme. ORF prediction by GeneMark also agrees with the upstream TIS. A longer bll2380 protein is also annotated in B. japonicum USDA6T and B. sp.23321.
Fig. 4.
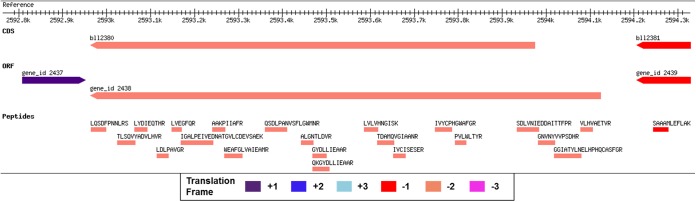
Gene model correction discovered by proteogenomics. Four novel peptides map upstream to annotated gene suggesting an upstream TIS. Four different tracks are shown. Reference track shows genomic co-ordinates. CDS track represent annotated protein coding genes. ORF track shows ORF predictions by GeneMark algorithm. Peptide track shows all the identified peptides with their respective sequences. Different bar colors represent different reading frames.
In cases where ORF predictions did not support gene model changes, we applied sequence similarity searches. One striking case is identified for NolA (bll2019), a transcriptional regulator protein which is a key member involved in nodulation in the host plant during symbiosis. The NolA protein is identified with six peptides. One of the peptides, IGELAEATGVTVR is identified in all nine samples and it uniquely maps upstream to the current annotation of the NolA TIS. All four gene predictions support the existing annotation of translational start site but do not cover the novel peptide. However, a longer protein is reported for some close strains by sequence similarity analysis (Fig. 5). Even for B. japonicum USDA110, a longer NolA protein is suggested in few studies (42, 43). Interestingly, the NolA locus of B. japonicum has been shown to code for three distinct proteins, varying in their TIS (44). The peptide is covered in the longest protein isoform from NolA locus and is a part of DNA binding domain in the protein's N terminus. The observation of the novel peptide in all nine samples suggests the longest protein isoform from NolA locus to be expressed during symbiosis with legume plants.
Fig. 5.
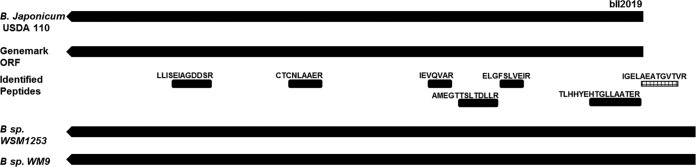
Gene model change for NolA gene (bll2019). Peptide IGELAEATGVTVR is identified upstream to current annotation of NolA gene on complement strand. GeneMark ORF prediction does not include the novel peptide. However, longer ortholog proteins are annotated for some closely related strains.
TIS Confirmation
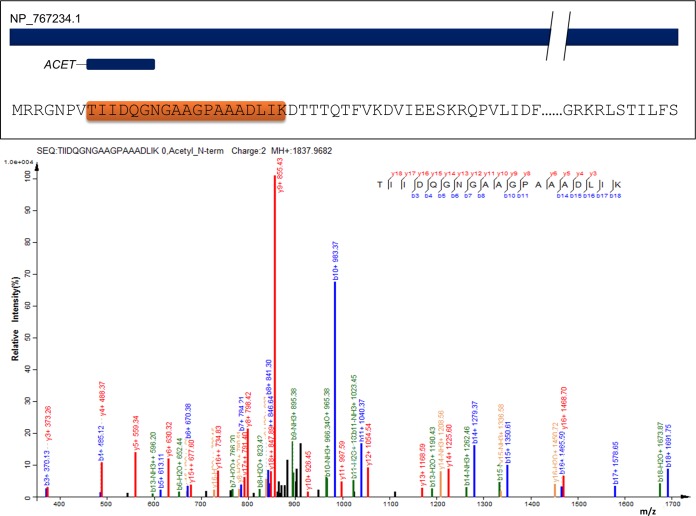
We analyzed protein translation initiation site by probing N-terminal specific modifications. Formylation happens on the initiator methionine and N-terminal acetylation on second amino acid after initiator methionine is cleaved. These modifications directly mark the TIS for a protein coding gene. Based on peptides identified at ≤1% FDR, and their upstream codon in the genome we could confirm the annotated TIS for 15 genes. Annotated peptide spectrum matches of selected N-acetylated peptides are provided in supplementary File S7. Additionally, we corrected TIS for one gene where N-terminally modified peptides did not agree with the currently annotated start site. Fig. 6 depicts an N-terminally acetylated peptide and a representative PSM, which helped in correcting TIS for the locus blr0594 (NP_767234.1), a thioredoxin protein. Peptide TIIDQGNGAAGPAAADLIK is identified with N-terminal acetylation. The codon preceding the N-terminally acetylated Threonine is GTG which is known to code for initiator methionine in genomes with high GC content. Based on the acetylated peptide we corrected the TIS for blr0594 locus. The newly assigned TIS is downstream to the annotated TIS for this locus. The orthologous protein sequences in related Bradyrhizobium genomes also agree with the newly assigned TIS for blr0594.
Fig. 6.
TIS correction discovered by N-acetylated peptides. Peptide TIIDQGNGAAGPAAADLIK and its peptide spectral match. Blue colored peaks are matched b-ion series. Red colored peaks are matched y-ion series. Black colored peaks are unassigned experimental peaks.
Proteogenomic Analysis of Shigella flexneri
We also applied GenoSuite on Shigella flexneri 2a str. 2457T data (Pride Accession 18992–18999). S. flexneri is a human pathogen which causes dysentery and diarrhea. We discovered 28 previously un-annotated protein coding genes and corrected annotation for 22 genes, which shows rapid proteogenomic analyses of another prokaryotic genome (supplementary File S8). No proteogenomic study for this bacterium has been reported so far. Novel and better annotations can lead to better understanding of pathogenesis by Shigella flexneri.
DISCUSSION
GenoSuite tool described here provides a simple and effective informatics framework for proteogenomic analyses from prokaryotes. To the best of our knowledge, there are no readily available tools dedicated for prokaryotic proteogenomic analyses. For instance, GAPP (45) is specific to human annotation and no longer actively developed. PepLine (46) uses a de novo tag based approach to detect peptides in a genome, which is usually more error prone than database searches. Integration of multiple algorithms in GenoSuite improves both sensitivity and specificity. Implementing FDScore method on different algorithms before integrating their outputs and calculating FDR at PSM and protein levels makes GenoSuite a robust statistical pipeline. It is currently designed to discover simple gene models found in prokaryotes.
As a proof of principle, we have used GenoSuite to analyze Bradyrhizobium japonicum USDA110 genome for discovering novel protein coding genes. B. japonicum USDA110 is an agriculturally important model organism to study symbiotic nitrogen fixation in legumes. A comprehensive annotation is a key prerequisite for such studies but its genome annotation is far from complete. Because only a handful of related genomes are sequenced, the annotation of protein coding genes could not take advantage of comparative genomics. GenoSuite identified large number of new proteins coding genes in B. japonicum USDA110. A significant number of these NPCRs are unique to this bacterium whereas others have orthology in two closely related genomes, namely B. japonicum USDA6T and B. sp.S23321. The exclusivity of these genes to one organism or to a small group of organisms makes them difficult to annotate by methods other than proteogenomics. Most of the NPCRs in our analysis are short in length (<200 aa). Previous proteogenomic studies in other organisms have also identified mostly short novel proteins (5, 6) and have speculated that short length of proteins could be a reason for missed annotation. Although Kaneko et al. (17) considered all ORF predictions of length 150 bp or above for gene prediction, many short genes within the acceptable length range (>150 bp) were left un-annotated in B. japonicum USDA110 genome. It is generally believed that genomes with high GC% suffer with over-prediction of short proteins (29, 47); our data suggests the opposite for B. japonicum genome, which also has high GC (64.1%). As NPCRs identified in proteogenomics studies are experimentally discovered, these can be a valuable resource for improving the existing gene prediction algorithms or development of new algorithms for predicting short length proteins. Additionally, by applying ortho-proteogenomics, we identified three new genes in the B. japonicum USDA6T genome. This indicates that although the computational gene prediction methods have improved, they still miss some protein coding genes and it emphasizes the use of proteogenomics as a powerful solution to such issues in genome annotation.
We also used novel peptides in the discovery of novel operons or correction of known operons. An operon is predicted when genes are on the same strand and the gap between them is not more than 50–60 bp. Many novel genes or N-terminal extension to existing genes reduce this gap and can correct operon models. In addition, it is a prerequisite that all proteins in new operon are identified. We discovered 11 operon models which could not be annotated based on initial annotations of protein coding genes. An example is an operon with six proteins related to Cytochrome P450 system. This operon is highly active in anaerobic conditions within symbiotic bacteroids and is crucial for normal nitrogen reduction within legume host. Christie-Oleza et al. also reported operons in Ruegeria pomeroyi by proteogenomic analysis (10). Correction and unveiling of operonic structures can aid in quick annotation as well as provide functional insights into the underlying biology of the organism.
Incorrect assignment of TIS is probably the most frequent error of genome annotation. We used fully tryptic peptides to correct such errors. We found changes in 48 existing gene models in B. japonicum genome. Most of the errors in TIS assignment were associated with TTG start codon. ATG and GTG start codons are preferred over TTG by gene prediction algorithms which may have resulted in incorrect assignment of TIS in observed cases. Protein sequences from these loci are significantly changed and may contribute to newer structural and functional features as shown in NolA gene where N-terminal extension provides additional DNA binding domain probably involved in a regulatory role. Protein N-terminal modifications can also be used to probe TIS. All TIS confirmations in our study are based on N-terminally acetylated peptides. N-Acetylation for blr0594 gene revealed that TIS is located downstream to the currently annotated site. N-terminal acetylation is believed to be rare in bacteria and only few proteins, mostly ribosomal, are known to be acetylated (48, 49). However, we observed diverse classes of proteins as N-acetylated in B. japonicum USDA110 which agrees well with a recent study by Bonissone S. et al. (50), which showed N-acetylation as a widespread protein modification among bacteria.
To establish GenoSuite's versatility and applicability in discovering novel translations, it was also applied on a recent Shigella flexneri 2a str. 2457T data where we discovered previously un-annotated genes and corrected annotation for several genes. This demonstrates rapid and automated proteogenomic analyses of prokaryotic genomes using GenoSuite. GenoSuite is a highly effective, easily configurable analysis suite for prokaryotic proteogenomics.
Supplementary Material
Acknowledgments
We thank Anupam Kumar Mondal for helpful discussions and insightful comments while proof-reading the manuscript.
Footnotes
* DK is supported by Department of Science and Technology-INSPIRE Senior Research Fellowship. AKY is supported by Council of Scientific and Industrial Research (India), Senior Research Fellowship. SHN is supported by UQ Postdoctoral Research Fellowship. SMG is supported by National Health and Medical Research Council (NHMRC) Principal Research Fellowship. DD acknowledges CSIR In silico Biology project (CMM-0017) and Genesis project (BSC-0121) for compute infrastructure and publication charges.
 This article contains supplemental Figs. S1 to S2, Tables S1 to S2 and Files S1 to S8.
This article contains supplemental Figs. S1 to S2, Tables S1 to S2 and Files S1 to S8.
1 The abbreviations used are:
- TIS
- Translation Initiation Site
- FDR
- False Discovery Rate
- PSM
- Peptide Spectrum Match
- NPCRs
- Novel Protein Coding Regions
- ORF
- Open Reading Frame.
REFERENCES
- 1. Gallien S., Perrodou E., Carapito C., Deshayes C., Reyrat J. M., Van, Dorsselaer A., Poch O., Schaeffer C., Lecompte O. (2009) Ortho-proteogenomics: multiple proteomes investigation through orthology and a new MS-based protocol. Genome Res. 19, 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aivaliotis M., Gevaert K., Falb M., Tebbe A., Konstantinidis K., Bisle B., Klein C., Martens L., Staes A., Timmerman E., Van, Damme J., Siedler F., Pfeiffer F., Vandekerckhove J., Oesterhelt D. (2007) Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J. Proteome Res. 6, 2195–2204 [DOI] [PubMed] [Google Scholar]
- 3. Lamontagne J., Béland M., Forest A., Côté-Martin A., Nassif N., Tomaki F., Moriyon I., Moreno E., Paramithiotis E. (2010) Proteomics-based confirmation of protein expression and correction of annotation errors in the Brucella abortus genome. BMC. Genomics 11, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gupta N., Benhamida J., Bhargava V., Goodman D., Kain E., Kerman I., Nguyen N., Ollikainen N., Rodriguez J., Wang J., Lipton M. S., Romine M., Bafna V., Smith R. D., Pevzner P. A. (2008) Comparative proteogenomics: combining mass spectrometry and comparative genomics to analyze multiple genomes. Genome Res. 18, 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kelkar D. S., Kumar D., Kumar P., Balakrishnan L., Muthusamy B., Yadav A. K., Shrivastava P., Marimuthu A., Anand S., Sundaram H., Kingsbury R., Harsha H. C., Nair B., Prasad T. S., Chauhan D. S., Katoch K., Katoch V. M., Kumar P., Chaerkady R., Ramachandran S., Dash D., Pandey A. (2011) Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol. Cell Proteomics 10, M111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Venter E., Smith R. D., Payne S. H. (2011) Proteogenomic analysis of bacteria and archaea: a 46 organism case study. PLoS. One. 6, e27587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yates J. R., 3rd, Eng J. K., McCormack A. L. (1995) Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal. Chem. 67, 3202–3210 [DOI] [PubMed] [Google Scholar]
- 8. Jaffe J. D., Berg H. C., Church G. M. (2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4, 59–77 [DOI] [PubMed] [Google Scholar]
- 9. Baudet M., Ortet P., Gaillard J. C., Fernandez B., Guérin P., Enjalbal C., Subra G., de G. A., Barakat M., Dedieu A., Armengaud J. (2010) Proteomics-based refinement of Deinococcus deserti genome annotation reveals an unwonted use of non-canonical translation initiation codons. Mol. Cell Proteomics 9, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Christie-Oleza J. A., Miotello G., Armengaud J. (2012) High-throughput proteogenomics of Ruegeria pomeroyi: seeding a better genomic annotation for the whole marine Roseobacter clade. BMC. Genomics 13, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castellana N. E., Payne S. H., Shen Z., Stanke M., Bafna V., Briggs S. P. (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U.S.A. 105, 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chaerkady R., Kelkar D. S., Muthusamy B., Kandasamy K., Dwivedi S. B., Sahasrabuddhe N. A., Kim M. S., Renuse S., Pinto S. M., Sharma R., Pawar H., Sekhar N. R., Mohanty A. K., Getnet D., Yang Y., Zhong J., Dash A. P., MacCallum R. M., Delanghe B., Mlambo G., Kumar A., Keshava Prasad T. S., Okulate M., Kumar N., Pandey A. (2011) A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry. Genome Res. 21, 1872–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhong Y., Chang X., Cao X. J., Zhang Y., Zheng H., Zhu Y., Cai C., Cui Z., Zhang Y., Li Y. Y., Jiang X. G., Zhao G. P., Wang S., Li Y., Zeng R., Li X., Guo X. K. (2011) Comparative proteogenomic analysis of the Leptospira interrogans virulence-attenuated strain IPAV against the pathogenic strain 56601. Cell Res. 21, 1210–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Castellana N., Bafna V. (2010) Proteogenomics to discover the full coding content of genomes: a computational perspective. J. Proteomics 73, 2124–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu W., Taylor J. A., Davis M. T., Bonilla L. E., Lee K. A., Auger P. L., Farnsworth C. C., Welcher A. A., Patterson S. D. (2010) Maximizing the sensitivity and reliability of peptide identification in large-scale proteomic experiments by harnessing multiple search engines. Proteomics 10, 1172–1189 [DOI] [PubMed] [Google Scholar]
- 16. Yadav A. K., Bhardwaj G., Basak T., Kumar D., Ahmad S., Priyadarshini R., Singh A. K., Dash D., Sengupta S. (2011) A systematic analysis of eluted fraction of plasma post immunoaffinity depletion: implications in biomarker discovery. PLoS. One 6, e24442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaneko T., Nakamura Y., Sato S., Minamisawa K., Uchiumi T., Sasamoto S., Watanabe A., Idesawa K., Iriguchi M., Kawashima K., Kohara M., Matsumoto M., Shimpo S., Tsuruoka H., Wada T., Yamada M., Tabata S. (2002) Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9, 189–197 [DOI] [PubMed] [Google Scholar]
- 18. Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L. (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koch M., Delmotte N., Rehrauer H., Vorholt J. A., Pessi G., Hennecke H. (2010) Rhizobial adaptation to hosts, a new facet in the legume root-nodule symbiosis. Mol. Plant Microbe Interact. 23, 784–790 [DOI] [PubMed] [Google Scholar]
- 20. Delmotte N., Ahrens C. H., Knief C., Qeli E., Koch M., Fischer H. M., Vorholt J. A., Hennecke H., Pessi G. (2010) An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 10, 1391–1400 [DOI] [PubMed] [Google Scholar]
- 21. Vizcaino J. A., Côté R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., Hermjakob H., Martens L. (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 38, D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yadav A. K., Kumar D., Dash D. (2011) MassWiz: a novel scoring algorithm with target-decoy based analysis pipeline for tandem mass spectrometry. J. Proteome. Res. 10, 2154–2160 [DOI] [PubMed] [Google Scholar]
- 23. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome. Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 24. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 25. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 26. Jones A. R., Siepen J. A., Hubbard S. J., Paton N. W. (2009) Improving sensitivity in proteome studies by analysis of false discovery rates for multiple search engines. Proteomics 9, 1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Käll L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome. Res. 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 28. Lukashin A. V., Borodovsky M. (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26, 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hyatt D., Chen G. L., Locascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC. Bioinformatics 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(2010) FGENESB: Bacterial Operon and Gene Prediction, In: http://linux1.softberry.com/berry.phtml?topic=fgenesb&group=programs&subgroup=gfindb
- 31. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290-D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCarthy F. M., Wang N., Magee G. B., Nanduri B., Lawrence M. L., Camon E. B., Barrell D. G., Hill D. P., Dolan M. E., Williams W. P., Luthe D. S., Bridges S. M., Burgess S. C. (2006) AgBase: a functional genomics resource for agriculture. BMC. Genomics 7, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., Formsma K., Gerdes S., Glass E. M., Kubal M., Meyer F., Olsen G. J., Olson R., Osterman A. L., Overbeek R. A., McNeil L. K., Paarmann D., Paczian T., Parrello B., Pusch G. D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. (2008) The RAST Server: rapid annotations using subsystems technology. BMC. Genomics 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Käll L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 35. Brosch M., Yu L., Hubbard T., Choudhary J. (2009) Accurate and sensitive peptide identification with Mascot Percolator. J. Proteome. Res. 8, 3176–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yadav A. K., Kumar D., Dash D. (2012) Learning from decoys to improve the sensitivity and specificity of proteomics database search results. PLoS. One 7, e50651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wedge D. C., Krishna R., Blackhurst P., Siepen J. A., Jones A. R., Hubbard S. J. (2011) FDRAnalysis: a tool for the integrated analysis of tandem mass spectrometry identification results from multiple search engines. J. Proteome. Res. 10, 2088–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hauser F., Pessi G., Friberg M., Weber C., Rusca N., Lindemann A., Fischer H. M., Hennecke H. (2007) Dissection of the Bradyrhizobium japonicum NifA+sigma54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol. Genet. Genomics 278, 255–271 [DOI] [PubMed] [Google Scholar]
- 39. Pessi G., Ahrens C. H., Rehrauer H., Lindemann A., Hauser F., Fischer H. M., Hennecke H. (2007) Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant Microbe Interact. 20, 1353–1363 [DOI] [PubMed] [Google Scholar]
- 40. Kaneko T. (2011) Complete Genome Sequence of the Soybean Symbiont Bradyrhizobium japonicum Strain USDA6T. Genes 2, 763–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okubo T., Tsukui T., Maita H., Okamoto S., Oshima K., Fujisawa T., Saito A., Futamata H., Hattori R., Shimomura Y., Haruta S., Morimoto S., Wang Y., Sakai Y., Hattori M., Aizawa S., Nagashima K. V., Masuda S., Hattori T., Yamashita A., Bao Z., Hayatsu M., Kajiya-Kanegae H., Yoshinaga I., Sakamoto K., Toyota K., Nakao M., Kohara M., Anda M., Niwa R., Jung-Hwan P., Sameshima-Saito R., Tokuda S., Yamamoto S., Yamamoto S., Yokoyama T., Akutsu T., Nakamura Y., Nakahira-Yanaka Y., Takada H. Y., Hirakawa H., Mitsui H., Terasawa K., Itakura M., Sato S., Ikeda-Ohtsubo W., Sakakura N., Kaminuma E., Minamisawa K. (2012) Complete Genome Sequence of Bradyrhizobium sp. S23321: Insights into Symbiosis Evolution in Soil Oligotrophs. Microbes. Environ. 27, 306–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Göttfert M., Röthlisberger S., Kündig C., Beck C., Marty R., Hennecke H. (2001) Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183, 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sadowsky M. J., Cregan P. B., Gottfert M., Sharma A., Gerhold D., Rodriguez-Quinones F., Keyser H. H., Hennecke H., Stacey G. (1991) The Bradyrhizobium japonicum nolA gene and its involvement in the genotype-specific nodulation of soybeans. Proc. Natl. Acad. Sci. U.S.A. 88, 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Loh J., Stacey M. G., Sadowsky M. J., Stacey G. (1999) The Bradyrhizobium japonicum nolA gene encodes three functionally distinct proteins. J. Bacteriol. 181, 1544–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shadforth I., Xu W., Crowther D., Bessant C. (2006) GAPP: a fully automated software for the confident identification of human peptides from tandem mass spectra. J. Proteome. Res. 5, 2849–2852 [DOI] [PubMed] [Google Scholar]
- 46. Ferro M., Tardif M., Reguer E., Cahuzac R., Bruley C., Vermat T., Nugues E., Vigouroux M., Vandenbrouck Y., Garin J., Viari A. (2008) PepLine: a software pipeline for high-throughput direct mapping of tandem mass spectrometry data on genomic sequences. J. Proteome. Res. 7, 1873–1883 [DOI] [PubMed] [Google Scholar]
- 47. Fukuchi S., Nishikawa K. (2004) Estimation of the number of authentic orphan genes in bacterial genomes. DNA Res. 11, 219–231, 311–313 [PubMed] [Google Scholar]
- 48. Soppa J. (2010) Protein acetylation in archaea, bacteria, and eukaryotes. Archaea. 2010, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu L. I., Lima B. P., Wolfe A. J. (2010) Bacterial protein acetylation: the dawning of a new age. Mol. Microbiol. 77, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bonissone S., Gupta N., Romine M., Bradshaw R. A., Pevzner P. A. (2012) N-terminal protein processing: A comparative proteogenomic analysis. Mol. Cell Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.