Fig. 3.
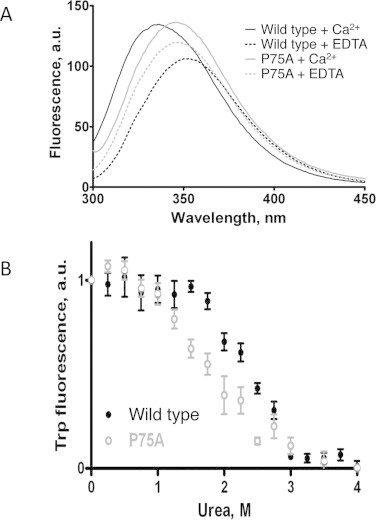
Elimination of proline from the C-terminus of ScaADoc (P75A mutant) effects folding and stability of the protein determined by intrinsic tryptophan fluorescence. (A) Structural changes upon Ca2+-binding to R. flavefaciens ScaADoc in tryptophan fluorescence wavelength scan. In the wild-type protein a blue fluorescence shift is observed as a result of Ca2+-induced folding. In the P75A mutant no change in hydrophobicity of the tryptophan environment occurred upon folding. Peaks of tryptophan fluorescence: 351 nm, wild type + EDTA (- - -); 335 nm, wild type + Ca2+ (—); 347 nm, P75A + EDTA ( ); 346 nm, P75A + Ca2+ (
); 346 nm, P75A + Ca2+ ( ). (B) Decreased stability of the P75A mutant R. flavefaciens ScaADoc. Assay of stability under conditions of increasing urea concentrations. Urea at a concentration of 1.8 M causes unfolding of half of the mutant ScaADoc constructs (○) versus 2.5 M for the wild-type ScaADoc (•).
). (B) Decreased stability of the P75A mutant R. flavefaciens ScaADoc. Assay of stability under conditions of increasing urea concentrations. Urea at a concentration of 1.8 M causes unfolding of half of the mutant ScaADoc constructs (○) versus 2.5 M for the wild-type ScaADoc (•).
