Figure 3. Patient BAB3027 breakpoint junction mutational load.
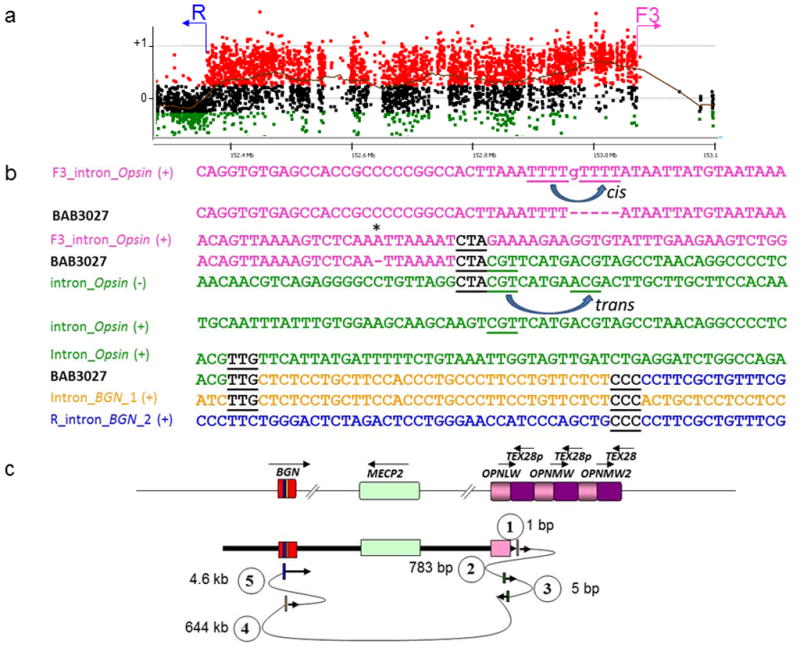
Patient BAB3027 presented at least three mutations at and flanking the CGR breakpoint junctions: a frameshift before the breakpoint junction, and multiple template-switch events. (a) BAB3027 aCGH result and approximate location of the primers (F and R) used to obtain patient specific breakpoint junctions. (b) Breakpoint junction sequence is aligned to the proximal and distal genomic references and color-matched. Strand of alignment (+ or −) is indicated in parenthesis. Microhomology at the breakpoint is indicated by black bold underlined letters. Dashed lines represent nucleotides that did not align to the reference sequence; asterisks indicate frameshifts flanking the breakpoint junction. Misalignment and re-annealing of short repeats present in the primer strand and template strand in cis can produce deletion in the newly synthetized strand (forward slippage) or insertion (backward slippage) 42. In addition, misalignment and re-annealing in trans would produce small inversion at the junctions 41. (c) Representation of the genomic structure for the reference genome (top) and for the surmised genomic structure (bottom), showing predicted order, origins, and relative orientations of duplicated sequences. Arrows show orientation of DNA sequence relative to the positive strand; filled arrows with circled numbers below represent a template switch that resulted in deletion or insertion of segments. Distance between the template switches are shown in bp or kb. The last arrow signifies resumption of replication on the original template which produced the CGR identified by aCGH.
