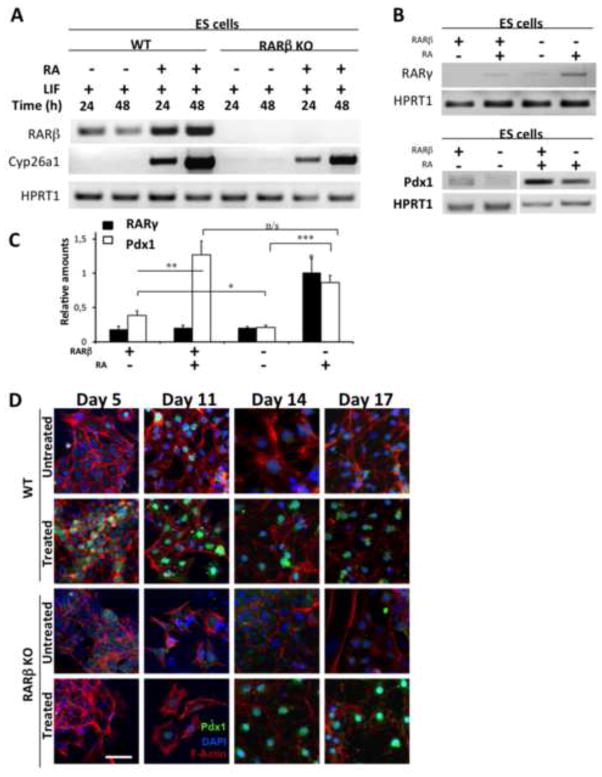
Figure 2. Impact of RARβ deletion on Pdx1 protein expression.
(A) RT-PCR analysis confirming the lack of RARβ transcripts in RARβKO ES cells. Analysis of Cyp26a1, a RA-responsive gene, demonstrates the presence of RA signaling activity via other receptors in RARβKO cells. HPRT1 was used as a loading control. (B) Representative RT-PCR analysis of Pdx1 and RARγ2 expression in WT and RARβ KO ES cells, control untreated (NT) and RA treated (1 μM, 48 h). (C) Relative amounts of Pdx1 and RARγ2, normalized to HPRT1 levels, are shown in the histogram (n=3; *: p≤0.047; **: p=0.001; ***: p≤0.0001). (D) Indirect immunofluorescence staining for Pdx1 protein (green) in WT and RARβKO cells at 5, 11, 14, and 17 days in the absence (untreated) or in the presence (treated) of growth factors used in the differentiation protocol. Cells were counterstained using rhodamine-conjugated phalloidin, which binds to F-actin (red), and nuclei were stained with DAPI (blue) (Bars = 50 μm).