Abstract
The GLD-2 class of poly(A) polymerases regulate the timing of translation of stored transcripts by elongating the poly(A) tails of target mRNAs in the cytoplasm. WISPY is a GLD-2 enzyme that acts in the Drosophila female germline and is required for the completion of the egg-to-embryo transition. Though a handful of WISPY target mRNAs have been identified during both oogenesis and early embryogenesis, it was unknown whether WISP simply regulated a small pool of patterning or cell cycle genes, or whether, instead, cytoplasmic polyadenylation was widespread during this developmental transition. To identify the full range of WISPY targets, we carried out microarray analysis to look for maternal mRNAs whose poly(A) tails fail to elongate in the absence of WISP function. We examined the polyadenylated portion of the maternal transcriptome in both stage 14 (mature) oocytes and in early embryos that had completed egg activation. Our analysis shows that the poly(A) tails of thousands of maternal mRNAs fail to elongate in wisp-deficient oocytes and embryos. Furthermore, we have identified specific classes of genes that are highly regulated in this manner at each stage. Our study shows that cytoplasmic polyadenylation is a major regulatory mechanism during oocyte maturation and egg activation.
Keywords: egg-to-embryo transition, cytoplasmic polyadenylation, egg activation, GLD2 family poly(A) polymerase, oocyte maturation
Introduction
Most eukaryotic mRNAs are polyadenylated at their 3’ terminus in a nuclear process that is coupled to their transcription. The presence of a poly(A) tail on a transcript both enhances its cytoplasmic stability and promotes its translation. In some cell types, a subset of transcripts has been shown to undergo further poly(A) tail adjustment in the cytoplasm. Typically, these transcripts undergo poly(A) tail shortening, which attenuates their translation, yet they remain stably ‘stored’ in the cytoplasm. In response to the appropriate cellular signals, these transcripts can then be induced to undergo cytoplasmic poly(A) tail lengthening, enabling their subsequent translation. As such, cytoplasmic polyadenylation of stored transcripts allows for rapid protein production in the absence of de novo transcription, enabling a potent mechanism for regulating gene expression.
The mechanism by which cytoplasmic polyadenylation is controlled has been studied in Xenopus oocytes (Mendez and Richter, 2001). Here, transcripts designated for poly(A) tail adjustments in the cytoplasm are marked by two cis-elements in the 3’UTR that interact with regulatory protein complexes, which include a poly(A) polymerase (PAP) as well as a ribonuclease that shortens the poly(A) tail. Upon a signal to activate translation, phosphorylation events within the protein complex cause the ribonuclease to be released (Kim and Richter, 2006). The cytoplasmic PAP is then free to elongate the poly(A) tail of the transcript, thus promoting its translation.
Important cellular and developmental transitions such as those during germline development rely on cytoplasmic poly(A) tail adjustments. For example, during late oogenesis and early embryogenesis prior to the maternal-zygotic transition, the transcriptional machinery of the cell is largely silent. Growing oocytes accumulate a large pool of maternal RNA molecules whose translation is repressed after their production and then activated post-transcriptionally through cytoplasmic polyadenylation for subsequent developmental progression during oogenesis and embryogenesis (Tadros and Lipshitz, 2005). Cytoplasmic polyadenylation that occurs in the germline is often mediated by the GLD-2 family of cytoplasmic PAPs. GLD-2 homologs have been identified in many species, including worms, flies, mice and frogs (Benoit et al., 2008; Cui et al., 2008; Kwak et al., 2004; Nakanishi et al., 2006; Sartain et al., 2011). The GLD-2-type PAP in the Drosophila female germline is encoded by the wispy (wisp) gene (Benoit et al., 2008; Cui et al., 2008). In the absence of wisp function, several transcripts necessary for development do not undergo poly(A) tail lengthening and as a result these mRNAs fail to become translated in a wispy null mutant (Benoit et al., 2008).
A candidate gene approach was previously used to identify specific maternal mRNAs that are subject to WISP-dependent cytoplasmic poly(A) regulation during Drosophila oocyte/embryo development (Benoit et al., 2008; Cui et al., 2008). Those studies focused on cell cycle regulators and maternal transcripts necessary for embryonic development as important WISP targets for cytoplasmic polyadenylation. However, the extent to which this process controls genome-wide regulation remained unknown. Here we describe a microarray-based approach designed to identify the full subset of maternal mRNAs that are targeted for WISP-dependent cytoplasmic polyadenylation. Our results indicate that WISP-dependent cytoplasmic polyadenylation is a major mechanism that regulates a wide spectrum of maternal mRNAs in the Drosophila female germline.
Materials and Methods
Drosophila stocks and sample collection
Male flies carrying the wisp null allele, wisp41 (Cui et al., 2008) were crossed to Df(1)RA47/FM7c female flies. wisp41/FM7c (wild type control) or wisp41/Df (wisp-deficient) virgin female progeny were separated from males and aged on standard yeast/glucose medium until use. Stage 14 oocytes were hand dissected from 3- to 5-day-old virgin females in hypertonic isolation buffer (Page and Orr-Weaver, 1997), which does not activate eggs. Virgin females that were 3 to 4 days old were mated to wild-type Oregon-R males and early embryos were collected 0- to 1-hr post egg deposition.
Sample preparation and hybridizing of microarrays
For global transcript analysis, we prepared total RNA as well as poly(A)+-selected RNA for microarray analysis. For each sample, total RNA was extracted from ~2000 stage 14 oocytes or 0- to 1-hr embryos using the TRIzol reagent (Invitrogen, Carlsbad, CA). Polyadenylated RNA was then isolated from total RNA using the Oligotex mRNA isolation kit (Qiagen, Valencia, CA) according to the manufacturer’s instruction. To test the efficiency and size-cutoff of the Oligotex kit in our hands, we ran a test-sample using heterogeneous poly(A) RNA (GE LifeSciences), which we had fragmented into a range between <10 nucleotides and >500 nucleotides. We found that Oligotex efficiently selected for poly(A) tracts of ~40 nt or greater (Figure S1). Sample preparation and hybridizing of microarrays was done as described (Pleiss et al., 2007), using 40 µg of total RNA or 800 ng of poly(A)-selected mRNA for cDNA synthesis. The purified cDNA was divided into halves. One half was conjugated to Cy3 and the other to the Cy5 fluorescent dye (GE Healthcare, Piscataway, NJ) to generate dye-swap pairs. Dye conjugation was performed as described (Pleiss et al., 2007).
Drosophila oligonucleotide microarrays (Agilent Design ID 18972, Agilent, Foster City, CA) were hybridized with labeled cDNA as described (Pleiss et al., 2007). Four groups of comparisons were performed: WT vs. wisp total RNA from oocytes, WT vs. wisp total RNA from fertilized eggs, WT vs. wisp poly(A)-selected RNA from oocytes, WT vs. wisp poly(A)-selected RNA from fertilized eggs. Each comparison consisted of three independent RNA extractions and each experiment was done with dye-swap pairs as two technical replicates.
Image processing, data pre-processing and analysis
Microarray images were acquired and analyzed as described (Pleiss et al., 2007). The six different experimental replicates (three independent biological samples with dye-flipped technical replicates of each) comparing the total RNA samples of wild type and wisp mutant samples were processed largely according to standard procedures (Smyth and Speed, 2003). For each of the six microarrays, ‘within array’ normalization was accomplished using the LOESS implementation in the marray package in Bioconductor. Processed data were then filtered according to expression level, excluding those features whose A-values were lower than 7.25 or greater than 15, and the remaining data were subsequently analyzed using Significance Analysis of Microarrays (SAM) to identify those genes whose expression was significantly changed in the mutant sample. We initially used SAM to identify enriched genes with False Discovery Rate (FDR) set to zero. To increase the stringency of our list of targets, we then considered only that subset of SAM-identified genes whose enrichment was at least 2-fold in magnitude. Importantly, no ‘between array’ processing was used to compare the six total RNA microarrays to one another. However, because the poly(A)+ RNA samples required such processing (see below), we also separately processed the total RNA arrays in an equivalent fashion. The lists of SAM identified genes that were determined both with and without ‘between array’ processing was nearly identical.
Microarrays comparing poly(A)+ RNAs were processed differently. Our initial analyses of these data suggested that there was good correlation between each of the different experimental replicates, but that the dynamic range varied significantly between them (Figure S1). Because each of these samples was subject to poly(A)+ purification, the effectiveness of which cannot be easily assessed, the absolute level of ‘enrichment’ detected for any given transcript is likely to vary from one microarray to the next. To account for these different dynamic ranges, quantile normalization was used to adjust the data from each of the six experimental replicates (Smyth and Speed, 2003). Briefly, the data from each replicate experiment were ranked from highest to lowest enrichment. From these lists, an average value was determined for each of the ranked positions. The average values were then re-assigned to the individual transcripts from each experiment according to their ranks (Figure S2). The validity of this approach was empirically confirmed using PCR-based polyA tail assays, as described below and in the Results section. After quantile normalization, we used SAM to identify genes that showed either enrichment or depletion within the poly(A)+ pool of wisp RNAs. Lists of enriched or depleted genes were tested by the DAVID Functional Annotation Bioinformatics Microarray Analysis (http://david.abcc.ncifcrf.gov/) to reveal over-represented Gene Ontology (GO) groups of annotated genes (Dennis et al., 2003).
Quantitative PCR and poly(A) tail assay
Total RNA was prepared as described above. Synthesis of cDNA was done using the SuperScript II Reverse Transcriptase kit (Invitrogen) according to the manufacturer’s protocol. Quantitative real-time PCR was performed using an ABI Prism 7000 system (Applied Biosystems, Foster City, CA). Each reaction was prepared in a 25 µl mixture containing 5 µl cDNA template, 0.2 µM of each primer and 1X SYBR Green Super Mix (Applied Biosystems).
PCR-based polyA tail (PAT) assay (Salles et al., 1994) was performed as previously described (Cui et al., 2008). Briefly, we incubated total RNA from our samples with an excess of poly(dT)12, which we then ligated. To the 5’ end of this strand we then attached an oligo(dT)12 anchor, and then we reverse-transcribed. PCR was performed on the PAT cDNAs using a gene-specific primer and the oligo(dT)12-anchor to assess the length of the poly(A) tail of a specific mRNA. PCR products from the PAT assay were separated on 8% acrylamide gels.
Cross-linking and RNA immunoprecipitation
Early embryos (0- to 1-hr post deposition) were collected from Oregon-R P2 flies as previously described. These embryos were permeabilized and cross-linked by shaking at room temperature in a 1:3 mixture of 1.8% formaldehyde cross-linking solution [50 mM HEPES (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1.8% formaldehyde] and heptane. RNA immunoprecipitation was done using the method in (Keene et al., 2006) with modification. Cross-linked embryos were washed in 1 X PBST with 125 mM glycine and homogenized in the homogenization buffer [10 mM HEPES (pH 7.0), 100 mM KCl, 5 mM MgCl2, 0.5% Nonidet P-40 (NP-40, Sigma), 1 mM dithiothreitol (DTT), 100 U/ml RNasin RNase inhibitor (Promega), 2 mM vanadyl ribonucleoside complexes (VRC, Sigma), protease inhibitor cocktail (Roche)] at 4°C. Lysates were filtered using Miracloth (EMD Chemicals, Gibbstown, NJ) and then centrifuged at 1,500 g for 10 minutes at 4°C. Protein concentration of supernatants was adjusted to 2 mg/ml by diluting in the immunoprecipitation buffer as described (Keene et al., 2006).
Protein A Sepharose beads (Sigma) were washed in the immunoprecipitation buffer with gentle shaking and centrifugation for 30 sec at 1,500 g. Egg extracts were pre-cleared with 50 µl of beads for 1 hr at 4°C, centrifuged at 1,500 g for 30 sec. The supernatant containing 5 mg of total proteins was incubated with 10 µg of anti-WISP antibody (Cui et al., 2008) or pre-immune serum (as control) and 50 µl of clean beads at 4°C with gentle rotating for 16 hr. The beads were collected with centrifugation and washed four times with the immunoprecipitation buffer and then four times with the immunoprecipitation buffer supplied with 1 M urea.
Beads were incubated in the elution buffer [100 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% sodium dodecyl sulfate (SDS), 40 U/ml RNasin RNase inhibitor] for 10 min with vortexing at 37°C and centrifuged at 1,500 g for 30 sec. The supernatants were treated with proteinase K (Roche) for 1 hr at 42°C and incubated for 1 hr at 65°C to reverse the cross-links. RNA was purified using TRIzol as described above. The abundance of each transcript was determined using qPCR. The signal for RPL32, which is not a WISP target and binds to the beads non-specifically, was used to normalize the raw Ct values among different samples. The fold enrichment for each transcript was then calculated as the difference of the normalized Ct values between the immunoprecipitated and mock samples.
Results
The poly(A)+ transcriptome is altered in the absence of WISP function
To determine the global changes in transcript polyadenylation status resulting from loss of WISP function, we used a microarray based approach to compare wild type and wisp null mutant samples at two different developmental stages: dissected stage 14 (mature) oocytes or fertilized activated eggs collected at 0- to 1-hr post egg deposition (hereafter referred to as “early embryos”). In an effort to distinguish between a change in the total level of a given transcript versus a change in its polyadenylation state, microarrays were performed that compared between the mutant and wild type flies either the total cellular RNA populations, or only those RNAs containing long poly(A) tails. Although conceptually similar to a previously described approach (Novoa et al., 2010), we undertook a different strategy to isolate these different populations (see Methods and/or SI for complete details). To compare levels of total RNA, independent of poly(A) state, between wild type and wispy strains, total cellular RNA was isolated and converted into cDNA using short, random oligonucleotides as primers for reverse transcriptase. Because these random primers initiate cDNA synthesis across the body of the transcript, conversion of RNA into cDNA occurs independent of the poly(A) status of the parent transcript. To address poly(A) state of these transcripts, portions of the same total cellular RNA preps were subjected to poly(A)+ purification using Oligotex mRNA isolation columns. Here we expect mRNAs with long poly(A) tails to bind the column more efficiently than those with short poly(A) tails: in diagnostic tests we determined that mRNAs require a poly(A) tail of approximately 40 or more adenosine residues for poly(A)-based purification using this kit (Figure S1). Notably, we previously observed that poly(A) tail lengths of WISP targets can become shorter in wisp null mutants (Cui et al., 2008). Therefore, we reason that the large difference in poly(A) tail length of a given target transcript between mutant and wild-type samples will result in differences in binding efficiencies between samples, such that the columns will efficiently recover transcripts with long poly(A) tails in a wild-type sample, but will poorly recover the same targets with shorter tails in the wisp null mutant. Multiple biological and technical microarray replicates were performed for each approach, and Significance Analysis of Microarrays (SAM) (Tusher et al., 2001) was used to identify RNAs whose total abundance was different between strains. As an additional stringency filter, we present here only those RNAs that were identified by SAM and whose enrichment level was at least 2-fold.
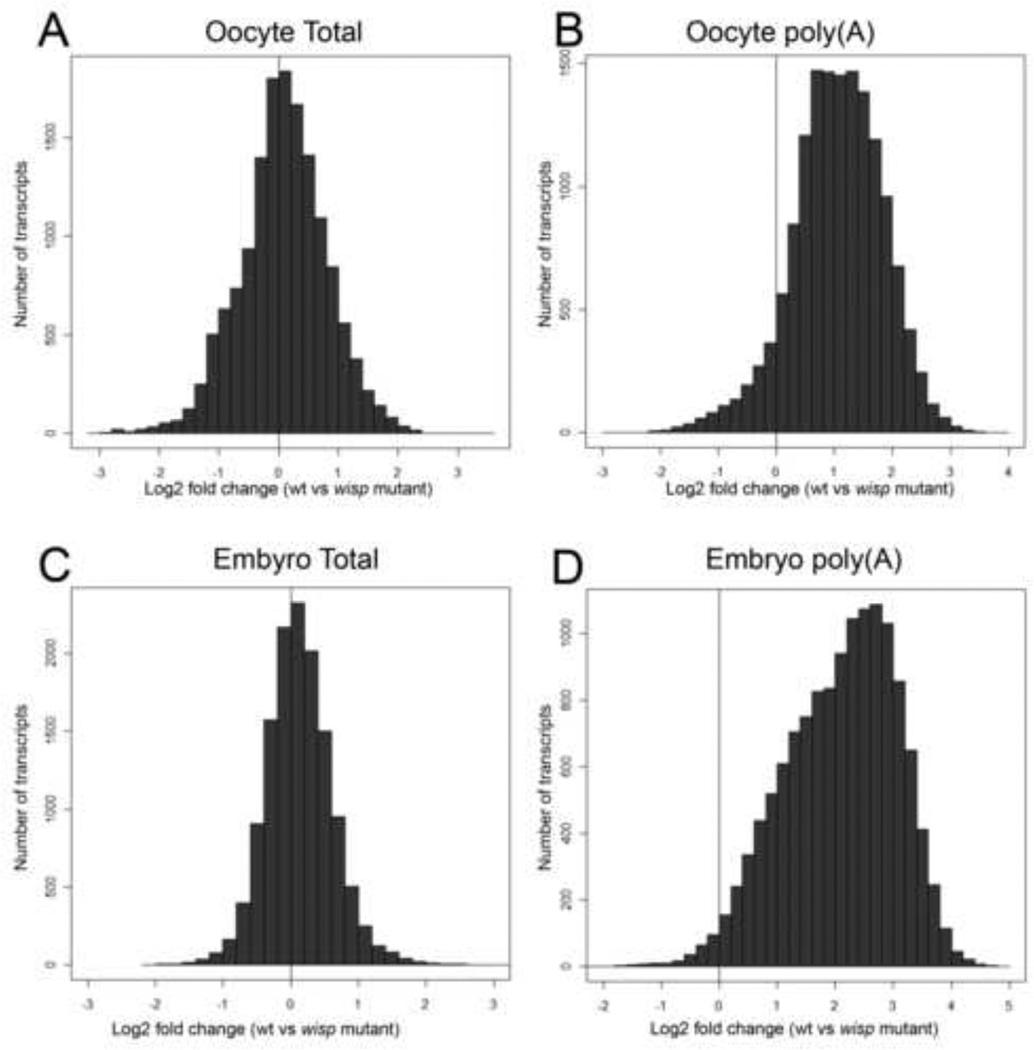
Relatively few changes in transcript abundance were detected between the total RNA samples at either developmental point (Fig. 1A, C). In the stage 14 oocytes, only 5 probes, corresponding to 2 genes, were enriched by more than two-fold between mutant and control samples (and none was more than three-fold enriched). By contrast, 560 (4%) probes were downregulated by more than two-fold (including 206 which were more than three-fold) (summarized in Table 1; total gene lists in Table S1). For the early embryos, the number of misregulated genes was lower still, with only 203 (2%) probes downregulated more than two-fold (only 32 of which were downregulated by more than three-fold) and no probes were significantly upregulated by more than two-fold.
Figure 1. Polyadenylated transcriptome is altered in the absence of WISP function.
Histograms of global comparisons between wild type and mutant show that for the majority of transcripts analyzed, total RNA abundance is not affected by the absence of WISP in stage 14 oocytes (A) or early embryos (C). However, within polyadenylated RNA populations, many transcripts are present in higher abundances in wild type than in wisp mutants in oocytes (B) and early embryos (D), indicating an overall downregulation of polyadenylated RNA in oocytes and embryos of wisp mutant mothers.
Table 1.
SAM analysis of genes that show ≥2-fold significant change in abundance.
| Higher in WT | Higher in wisp− | Array Total | ||||
|---|---|---|---|---|---|---|
| Probes | Genes | Probes | Genes | Probes | Genes | |
| Oocyte Total | 560 | 359 | 5 | 2 | 14886 | 6359 |
| Oocyte polyA | 5849 | 2613 | 2 | 2 | ||
| Embryo Total | 203 | 133 | 0 | 0 | 13208 | 5697 |
| Embryo polyA | 11567 | 4171 | 0 | 0 | ||
By contrast with our observations of the total RNA levels, the poly(A)+ RNA population from wild type was remarkably different from that of the wisp null mutant (Fig.1B, D). In stage 14 oocytes, a total of 5849 probe sets (out of 14886, or 39%) detected on the microarrays were down regulated by at least two-fold, with 3298 of these showing down-regulation of at least three-fold. The 5849 probe sets further collapsed into 2613 unique genes (Table 1, full list of genes in Table S1). Remarkably, early embryos showed even larger changes with 11567 probe sets (out of 13208, or 88%) detected on the microarrays showing down-regulation of at least twofold, and 9492 of these showing down-regulation of at least three-fold. The 11567 probe sets represented transcripts from 4171 genes (Table 1) (full list of genes in Table S1).
There was significant overlap between the probes downregulated among total RNA samples and the probes downregulated among poly(A)+ samples. In the oocyte dataset, where 560 probes are downreglated by 2-fold or more, 87% are also decreased in abundance in the poly(A)+ dataset. In the embryo dataset, all 203 probes downregulated amongst total RNA are also downregulated in the poly(A)+ dataset. For these probes, we cannot distinguish between the possibility that their depletion in the poly(A)+ pools is a result of shortened poly(A) length leading to their degradation, versus a simple decrease in their overall abundance. Nevertheless, these results are consistent with our general expectation that difference in total mRNA expression between the mutant and wild type should be similarly reflected in the poly(A)+ samples. By contrast, the remainder of the probes that are downregulated in the poly(A)+ pool did not show a significant difference in total mRNA expression between the wild type and mutant strains, strongly suggesting that their behavior in these poly(A)+ microarrays reflects a shortening of their poly(A) tails.
Among the 4171 putative WISP targets detected in early embryos, 2318 (56%) overlapped with the putative targets detected in stage 14 oocytes. These results are consistent with a model in which these RNAs have shorter poly(A) tails in wisp null mutants than controls during oogenesis, and the short poly(A) tails of these mRNAs are sustained in wisp mutants until after egg activation in the early embryos. However, we cannot rule out the possibility that they could be regulated by cytoplasmic polyadenylation during both oogenesis and egg activation. Some mRNAs such as the cyclin B transcript are known to undergo two rounds of polyadenylation (Benoit et al., 2005; Vardy and Orr-Weaver, 2007). It is possible that other genes could be regulated in the same way as cyclin B mRNA. The remaining 1853 probe sets were only detected as WISP regulated targets in early embryos, suggesting that their poly(A) tails undergo WISP-dependent elongation only upon egg activation.
Poly(A) tail length assays confirm the microarray results
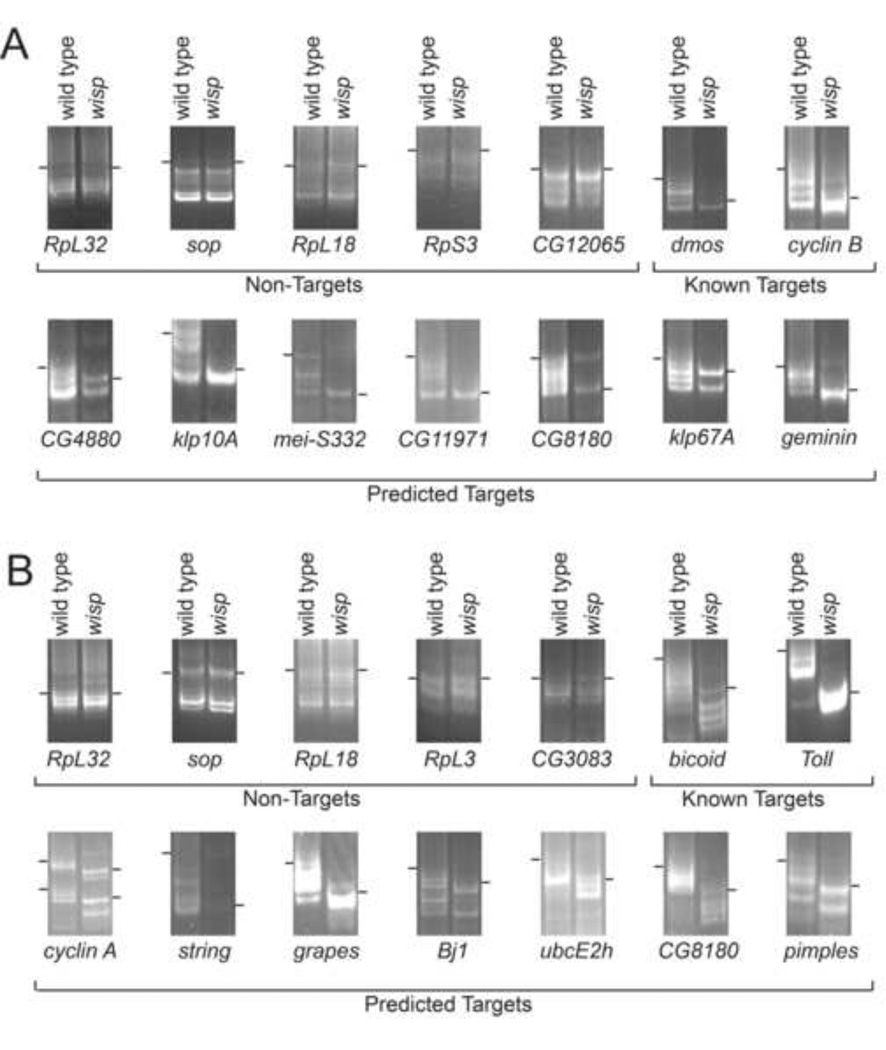
Because of the challenges associated with analyzing the poly(A)+ microarray data (see Materials and Methods), we sought to examine directly the poly(A) tail lengths of putative targets from each group using a PCR-based poly(A)-tail (PAT) test (Salles et al., 1994). We chose to test target sets that represented the full range of positive hits predicted by the microarray data, according to rank order of statistical significance as determined with SAM, and use these data to help establish the subset of high-confidence targets of WISP. For stage 14 oocytes, we chose nineteen putative WISP egg maturation targets identified above, ranging in rank from 6 to 4077 (Table 2): CG4880, CG11971, Klp10A, CG8180, klp67A, CDC6, sced, mei-S332, geminin, AurB, sse, cyclin A, HSF, sce, psq, mr, bifocal, eIF4G, and CG5316. Two previously known WISP-regulated RNAs, dmos and cyclin B (Benoit et al., 2008; Cui et al., 2008), appeared on our list (ranks 260 and 1083, respectively) and were included as positive controls. Similarly, in early embryos we chose twenty putative target mRNAs ranking in significance from 19 to 6551 (Table 2): pimples, punt, spinster, CG8485, costa, Bj1, CG5262, CG2921, CG33298, Pp1α-96A, ubcE2h, tango11, Dsor1, CG8180, grapes, string, CG10209, mei-p26, CG8370, and CG34398. Two previously known WISP-regulated RNAs, bicoid and Toll (Benoit et al., 2008; Cui et al., 2008), were also identified on our list (ranks 2293 and 3313, respectively) and included as positive controls. As shown in Figure 2 and Table 2, the poly(A) tail shortening of ~40 to ~120 nts was seen for all positive controls and the vast majority of the test mRNAs in the absence of WISP. Remarkably, the top 16 candidates in oocytes, representing the top ~3000 target transcripts, all showed shortened poly(A) tail length in wisp mutants by the PAT assay, suggesting a high true positive discovery rate for the microarray data. Similarly, 17 of the 20 candidates tested in early embryos confirmed the shortened tail phenotype, demonstrating that upwards of ~6000 transcripts have shortened poly(A) tails in early embryos of the wisp null mutant.
Table 2.
PAT assay validation of selected candidate target mRNAs.
| Oocyte | ||||
| Gene name | Transcript ID | Rank | Score(d) | PAT assay |
| CG4880 | CG4880-RA | 6 | 4.76 | + |
| CG11971 | CG11971-RA | 23 | 4.15 | + |
| Klp10A | CG1453-RE | 38 | 4.02 | + |
| CG8180 | CG8180-RA | 39 | 4.01 | + |
| klp67A | CG10923-RA | 40 | 4.01 | + |
| CDC6 | CG5971-RA | 83 | 3.82 | + |
| sced | CG3273-RA | 103 | 3.76 | + |
| mei-S332 | CG5303-RA | 170 | 3.59 | + |
| dmos* | CG8767-RA | 260 | 3.45 | + |
| geminin | CG3183-RA | 369 | 3.31 | + |
| AurB | CG6620-RA | 540 | 3.16 | + |
| sse | CG10583-RA | 709 | 3.05 | + |
| cyclin B* | CG3510-RA | 1083 | 2.84 | + |
| cyclin A | CG5940-RA | 1247 | 2.77 | + |
| HSF | CG5748-RD | 1557 | 2.66 | + |
| sce | CG5595-RA | 1896 | 2.56 | + |
| psq | CG2368-RI | 2968 | 2.28 | − |
| mr | CG3060-RA | 3225 | 2.21 | − |
| bifocal | CG1822-RB | 3899 | 2.06 | + |
| eIF4G | CG10811-RB | 4013 | 2.03 | + |
| CG5316 | CG5316-RB | 4077 | 2.02 | − |
| Embryo | ||||
| Gene name | Transcript ID | Rank | Score(d) | PAT assay |
| pimples | CG5052-RA | 19 | 4.18 | + |
| punt | CG7904-RA | 197 | 3.76 | + |
| spinster | CG8428-RB | 300 | 3.67 | − |
| CG8485 | CG8485-RD | 492 | 3.55 | + |
| costa | CG1708-RA | 600 | 3.48 | − |
| bj1 | CG10480-RA | 737 | 3.43 | + |
| CG5262 | CG5262-RA | 787 | 3.40 | + |
| CG2921 | CG2921-RA | 998 | 3.33 | + |
| CG33298 | CG33298-RB | 1087 | 3.30 | − |
| Pp1alpha-96A | CG6593-RA | 1376 | 3.21 | + |
| ubcE2h | CG2257-RA | 1569 | 3.16 | + |
| tango11 | CG30404-RB | 1674 | 3.13 | + |
| Dsor1 | CG15793-RA | 2146 | 3.03 | + |
| CG8180 | CG8180-RA | 2164 | 3.03 | + |
| bicoid* | CG1034-RE | 2293 | 3.00 | + |
| grapes | CG17161-RA | 2346 | 2.99 | + |
| string | CG1395-RA | 2477 | 2.96 | + |
| Toll* | CG5490-RA | 3313 | 2.78 | + |
| CG10209 | CG10209-RA | 3738 | 2.69 | + |
| cyclin A | CG5940-RA | 3767 | 2.68 | + |
| mei-p26 | CG12218-RA | 5702 | 2.29 | + |
| CG8370 | CG8730-RA | 6476 | 2.13 | + |
| CG34398 | CG34398-RC | 6551 | 2.11 | − |
previously identified WISP target
Shift in electrophoretic mobility due to WISP
No shift in electrophoretic mobility due to WISP
Figure 2. Poly(A) tails are shorter in the absence of WISP function.
Poly(A) tail length of mRNAs was assessed in stage 14 oocytes (A) or activated eggs (B) using gene-specific primers in PAT assays, including four ribosomal protein mRNAs and ten mRNAs. Bars indicate the longest products produced by PAT; their absolute sizes were determined relative to standards, as shown in Fig. S4. Although laddering occurred in some PAT assays, it is important to note that in every case the amplicon was always larger in the samples from controls, whose longer poly(A) tails can bind more d(T) units, than in samples from wispy mutants, where the tested RNAs’ poly(A) tails were shorter. “Predicted targets” are those predicted from our microarray data. “Known targets” and “non-targets” were identified previously as WISP targets or WISP independent (respectively) by Benoit et al. (2008) and Cui et al. (2008).
Due to the difficulty associated with amplifying the entire length of the poly(A) tail for a given transcript of interest, the PAT assay itself is subject to a high false negative discovery rate. For example, previously uncharacterized alternative 3’UTR isoforms have been discovered in C. elegans and likely exist in other model organisms (Jan et al., 2011). Such alternative isoforms could impede PCR amplification of 3’UTR and poly(A) tail regions, causing the transcript-specific primers of the PAT assay to either not bind (shorter isoforms) or to be too far upstream for the allowed extension time (longer isoforms). As such, it remains unclear whether the few candidates that failed to show decreased poly(A)+ tail lengths in the absence of WISP by PAT assay are, in fact, false positive discoveries from the microarray, or whether their true 3’UTRs are simply misannotated in the sequence databases, leading to unclear PAT assay results. However, when alternative poly(A) sites occurred within a short distance of one another, our PAT assays were able to detect polyadenylation of transcripts with either of the alternative sites (for example, cyclin A shows two distinct polyadenylation sites in PAT assays in early embryos, Figure 2B). In the oocyte set, all 16 of the tested candidates within the top ~3000 most enriched showed clear poly(A) tail shortening in the absence of WISP, after which the success rate dropped off (Table 2). For these data, we thus posit that the limit of the reliable microarray data is reached at or near the 3000 transcript mark. Interestingly, SAM revealed approximately 3000 transcripts that are upregulated by 3-fold or more among control poly(A)+ RNA populations, thus supporting inclusion of data within this limit. By contrast, three of the transcripts from the early embryo set failed to demonstrate poly(A) tail shortening in the absence of WISP by the PAT assay in spite of sometimes strong enrichment on the microarrays. However, there is a strong overall confirmation rate within the early embryo data set through the entire list of ~6000 transcripts. SAM revealed more than 8000 transcripts that are present at levels 3-fold more or higher in the poly(A)+ RNA population of controls compared to that of wisp mutants.
We noticed that in wisp deficient oocytes all the tested mRNAs retain a short poly(A) tail of ~10 to ~40 nt, depending on the specific mRNA. This observation has been reported previously for bicoid mRNA in wisp mutants (Cui et al., 2008). The residual tail likely results from the function of the nuclear poly(A) polymerase, encoded by the hiiragi gene, which is active in early oogenesis (Benoit et al., 2008).
qPCR also validates the shortened poly(A) tails in wisp mutants
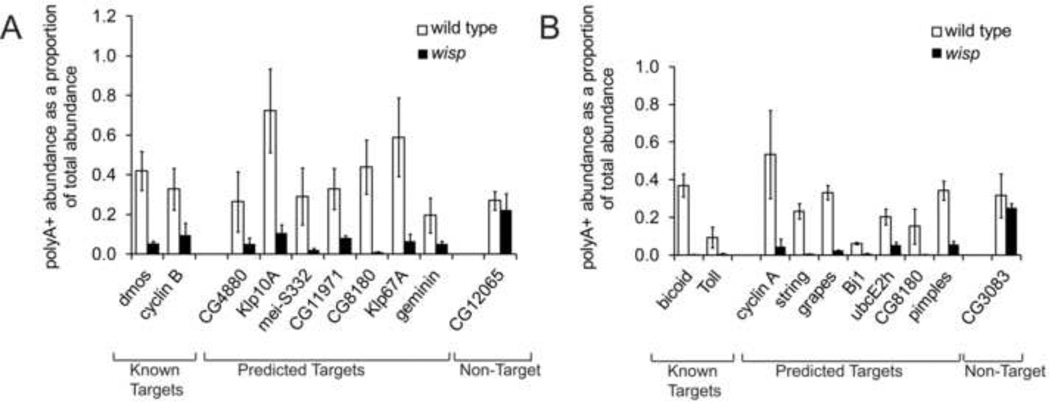
To further validate the microarray results, we used real-time quantitative PCR (qPCR) to quantify the RNA levels of selected candidates in both the total RNA and poly(A)+ pools. In stage 14 oocytes, seven mRNAs from the above list were chosen: CG4880, klp10A, mei-S232, CG11971, CG8180, klp67A, and geminin. CG12065 was used as a negative control. Two previously known WISP-dependent RNAs, dmos and cyclin B, were used as positive controls (Benoit et al., 2008; Cui et al., 2008). We used qPCR to assess the abundance of poly(A) RNA versus total RNA for these mRNAs in wild type and wisp oocytes and plotted the ratio of poly(A)+ RNA/total RNA for each transcript. As shown in Figure 3A, all seven putative targets and both positive controls, but not the negative control, showed a decreased ratio of poly(A)+ RNA/total RNA in the wisp mutant compared to wild type, suggesting that wisp null mutant affected the poly(A) tail length of these mRNAs.
Figure 3. Polyadenylated transcripts are depleted in the absence of WISP.
qPCR analysis measures the poly(A)+ RNA abundance, shown as a proportion of the total RNA abundance in stage 14 oocytes (A) or activated eggs (B). Results from wild type oocytes (unfilled bars) and from wisp deficient oocytes (filled bars) are shown for two biological replicates and one technical replicate. Error bars indicate standard deviation.
In early embryos, we tested seven candidates from the above list: cyclin A, string, grapes, Bj1, ubcE2h, CG8180, and pimples. CG3083 was used as a negative control. Two previously known WISP-regulated RNAs, bicoid and Toll were used as positive controls in the experiment. As shown in Figure 3B, all seven putative targets and two positive controls, but not the negative control, showed a decreased ratio of poly(A)+ RNA/total RNA in the wisp null mutant compared to wild type, suggesting that wisp mutant affected the polyadenylation of these mRNAs.
Functional relevance of genes regulated by WISP in stage 14 oocytes
We performed Gene Ontology (GO) annotation analysis to determine which functional groups of genes are over-represented in putative WISP targets in mature oocytes using DAVID (http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003). Only genes whose poly(A)+ transcript levels were greater by 2-fold or more in controls than in wisp mutants were included in this analysis. The total list of probes identified on the array for the mature oocyte poly(A)+ sample was used as the background sample. GO analysis revealed that “cell cycle” related genes made up the top category within Biological Process (P value=1.23E-22; Bonferroni-adjusted=2.84E-19) [for a full list of terms see Tables S2 (>2-fold), S3 (>3-fold), and S8]. These groups of WISP targets include genes that encode cell cycle regulators, such as cyclins (cyclin A, cyclin B, cyclin B3, cyclin D and cyclin E) and checkpoint regulators (wee and polo kinases). The enrichment of cell cycle related genes can be linked to the phenotypes observed in oocytes produced from wisp mutant mothers. One such example is klp3A, which encodes a kinesin-like protein that is required for pronuclear migration at the end of meiosis (Williams et al., 1997). Previously we observed that the female pronucleus does not migrate in wisp mutant (Cui et al., 2008). This phenotype could be due to the lack of translation of klp3A mRNA when its poly(A) tail fails to lengthen in the absence of WISP function.
Top categories in Molecular Function are “DNA binding” (P value =3.71E-17; Bonferroni-adjusted 3.16E-14) and “zinc ion binding” (P value =2.75E-11; Bonferroni-adjusted 2.35E-08) (Table S3). Emerging evidence suggests that zinc is critical for proper development of oocytes. During maturation, mouse oocytes accumulate over twenty billion zinc atoms, which are exocytosed in up to five rapid release events during egg activation (Kim et al., 2011). Zinc function is critical for progression of oocytes through the meiosis I–meiosis II transition in mouse oocytes (Bernhardt et al., 2011). In Drosophila oocytes, zinc-binding proteins are also enriched and are modulated by phosphorylation or dephosphorylation during egg activation (Krauchunas et al., 2012).
Using the KEGG pathway identifier in GO analysis, we found that members of the ubiquitination machinery were highly enriched among the WISP-regulated RNA groups in oocytes (P = 1.57E-07; Bonferroni-adjusted 1.38E-05) (Table S3). Among these are two E1 ubiquitin-activating enzymes, 14 E2 ubiquitin-conjugating enzymes, and most subunits of the two major multi subunit E3 ubiqitin ligase complexes, Cullin-Rbx and Anaphase-promoting complex (APC/C) (annotations from KEGG PATHWAY Database, http://www.genome.jp/kegg/pathway.html). Regulatory protein degradation plays important roles in oocyte development. In Xenopus oocytes, the E3 ubiquitin ligase APC/C degrades XErp1 protein, one component of the cytostatic factor, and this degradation causes the release of meiotic arrest (Wu and Kornbluth, 2008). The APC/C is also involved in the activation of meiotic progression in Drosophila oocytes. A mutation of the cortex gene, which encodes a female meiosis-specific activator of APC/C, results in the accumulation of Cyclin A protein and blocks meiotic progression (Pesin and Orr-Weaver, 2007). Polyadenylation of cortex mRNA occurs during oocyte maturation (Pesin and Orr-Weaver, 2007) and this polyadenylation depends on WISP function (Benoit et al., 2008). Our finding suggests that the ubiquitination machinery is probably activated as a consequence of cytoplasmic polyadenylation during late oogenesis, thus allowing cell cycle progression at egg maturation.
WISP-regulated RNAs are also associated with the “progesterone-mediated oocyte maturation” pathway (P = 1.16E-04; Bonferroni-adjusted 1.02E-2) (Table S3), a pathway that has been characterized previously in Xenopus (Belloc et al. 2008). In Xenopus, a signal from progesterone ultimately results in cytoplasmic polyadenylation and subsequent translation of stored mRNAs, which plays a central role in activating oocyte maturation in Xenopus (Belloc et al., 2008). Drosophila oocyte maturation does not involve progesterone, but the goal of oocyte maturation—release from one meiotic arrest point and progression to the next meiotic arrest point—remains conserved. Therefore, it is not surprising that some of the factors in the Xenopus progesterone-mediated oocyte maturation pathway, such as those that regulate cell cycle dynamics, would also be required during Drosophila egg maturation. Like the Xenopus oocyte maturation pathway involving MAPK and MOS, we have found that MAPK activity is high in Drosophila oocytes, and that mos mRNA is among WISP targets at egg maturation (Cui et al., 2008), further confirming conservation of key signaling events at egg maturation between Drosophila and Xenopus. In contrast to Xenopus, however, Drosophila MOS is not required for completion of meiosis (Ivanovska et al., 2004).
Genes that were identified as downregulated in the total RNA population (Table 1) are enriched for many of the same functional categories as those in the poly(A)+ population, including cell cycle and chromosome organization categories (Table S4); this is expected, since many transcripts that are downregulated in total RNA are shared in the poly(A)+ group.
Functional relevance of genes regulated by WISP in early embryos
We also searched for enriched functional groups among the genes that are WISP regulated in early embryos after egg activation. Enriched functional categories within Biological Process are indicative of cellular changes that occur during egg activation, including “reproductive cellular process” (P value =4.32E-10; Bonferroni-adjusted 1.36E-06), “protein amino acid phosphorylation” (P value =6.36E-07; Bonferroni-adjusted 2.00E-3), and “pattern specification process” (P value =7.23E-06; Bonferroni-adjusted 2.25E-2) [for a full list of terms see Tables S5 (>2-fold),S6 (>3-fold) and S9]. Top-ranking categories in Cellular Compartment suggest that chromosome-related proteins are enriched in WISP targets (P value =1.36E-14; Bonferroni-adjusted 8.29E-12), which may reflect release from meiotic arrest and progression to embryonic mitotic cell cycles. Top categories in Molecular Function are “zinc ion binding proteins” (P value =7.16E-14; Bonferroni-adjusted 1.02E-10) and “protein kinase activity” (P value =2.41E-08; Bonferroni-adjusted 3.43E-05), which likely relate to the signaling events that initiate and coordinate the process of egg activation.
GO analysis identified the functional group “pattern specification process” (Table S6). This group includes WISP targets that are known to function in patterning the embryo, such as bicoid, Toll, torso, caudal, gurken, hunchback, nanos, oskar, pumilio, and trunk. Posttranscriptional regulation of axis patterning genes in Drosophila embryos has been documented, and some of these genes’ transcripts are already known to be regulated at the level of cytoplasmic polyadenylation. For example, translation of oskar mRNA in the embryo’s posterior is activated by cytoplasmic polyadenylation (Castagnetti and Ephrussi, 2003; Chang et al., 1999). Additionally, the maternally supplied hunchback mRNA poly(A) tail elongates to about 70 nucleotides in early embryos; this correlates with the increase of its translation (Wreden et al., 1997). Furthermore, changes in poly(A) tail length of bicoid has been described previously (Salles and Strickland, 1999) and attributed to WISP function (Benoit et al., 2008; Cui et al., 2008). Although some of these pattern formation genes are already known to be targets of regulatory polyadenylation, it was not known that others are also regulated in a poly(A)-dependent manner. Our GO analysis reveals that transcripts of pattern specification genes comprise a major class among the total number of WISP targets at egg activation.
Because some target transcripts identified in early embryos were also identified as WISP targets in mature oocytes, we additionally analyzed the subset of transcripts that are unique to the embryo array and were not identified as targets in the mature oocyte array. GO analysis indicates that transcripts from genes involved in embryogenesis and morphogenesis are enriched within this subset, but not to the level of our significance cut-off.
Functional relevance of RNAs that are not regulated by WISP
We also performed GO analysis on the sets of genes whose transcripts were not affected by the presence vs. absence of WISP in oocytes and early embryos. Genes involved in mitochondrial cellular respiration were highly represented in both oocyte and embryo sets (Table S7). The most highly enriched functional categories within Biological Process include “oxidation reduction” for oocytes (P value =2.98E-9; Bonferroni-adjusted 8.51E-6) and “oxidative phosphorylation” for early embryos (P value =7.68E-21; Bonferroni-adjusted 7.19E-18) (Table S7). Similarly, enriched functional categories within Cellular Compartment included “mitochondrial respiratory chain” for both oocyte (P value =2.16E-5; Bonferroni-adjusted 1.21E-2) and embryo (P value =1.62E-17; Bonferroni-adjusted 3.99E-15). Genes within these groups for both data sets include subunits of cytochrome C oxidase (CoIV, CoVa, CoVb, CoVIb, and CoVIIc) as well as energy metabolism enzymes such as aldehyde dehydrogenase (Aldh) and glutamate dehydrogenase (Gdh). These results indicate that while WISP regulates many cellular changes during egg maturation and egg activation, genes and transcripts governing basic cellular energy requirements are not affected in a WISP-dependent manner. The mitochondrially-encoded gene mt:ND2 was also found among the genes that are not WISP-regulated, indicating that transcripts from the mitochondrial genome likely remain unaffected.
Target RNAs can be co-immunoprecipitated with WISP
With such a large part of the maternal transcriptome showing poly(A) tail shortening in the wisp mutant, the question arises whether these transcripts are direct targets of WISP. Although GLD-2-type poly(A) polymerases do not have RNA binding domains (Wang et al., 2002) and they are believed not to bind RNA directly, GLD-2 proteins can be recruited to the target mRNA by forming complexes with certain RNA binding proteins (Barnard et al., 2004; Kim et al., 2009; Wang et al., 2002). It has also been shown that WISP protein and RNA are in the same ribonucleoprotein (RNP) complex in ovarian extracts (Benoit et al., 2008). It is likely that mRNAs directly regulated by WISP will also be found in such WISP-containing RNP complexes.
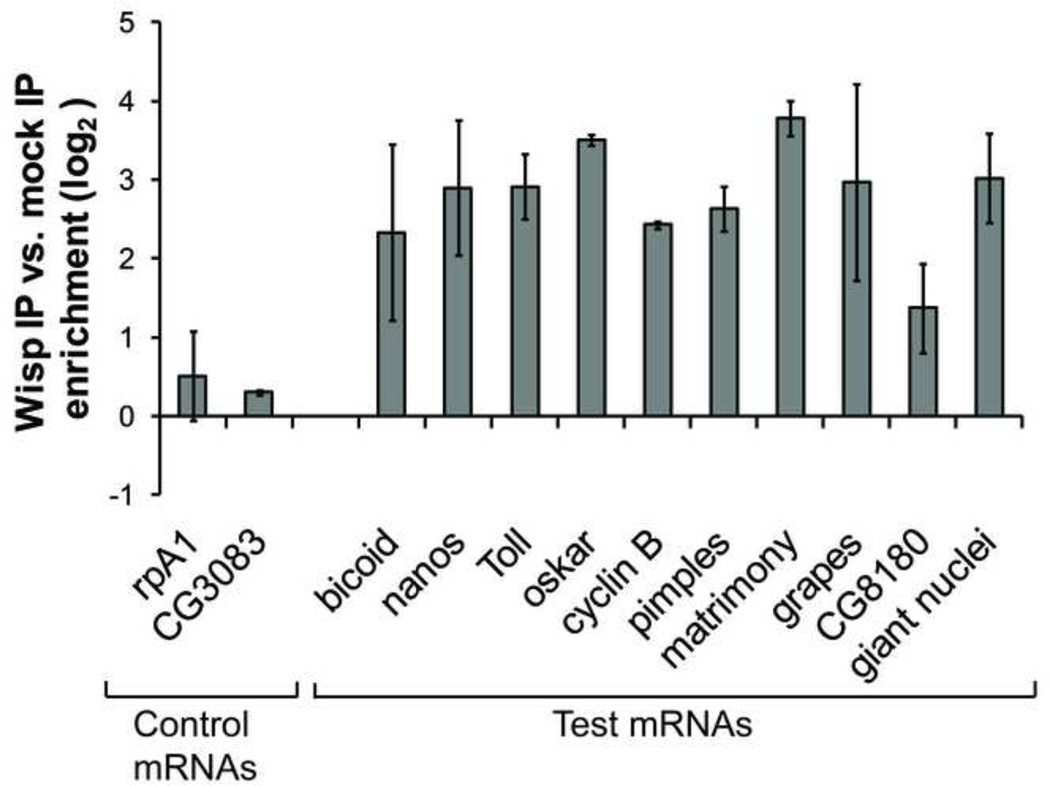
We used RNA immunoprecipitation (Keene et al., 2006) to determine whether a subset of putative WISP targets were associated with WISP-containing RNPs. Cytoplasmic extracts were made from wild type early embryos, and WISP-containing RNPs were immunoprecipitated from these extracts using an immobilized polyclonal anti-WISP antibody (Cui et al., 2008); pre-immune serum was used in parallel as a negative control. We used real-time quantitative RT-PCR to examine the enrichment of RNA that was immunoprecipitated with WISP. Test mRNAs included seven known targets of WISP (Benoit et al., 2008; Cui et al., 2008) as well as three targets identified in this microarray study and confirmed with PAT assays in Figure 3C. All of the ten test mRNAs were enriched by >2-fold in the WISP IP compared with the mock IP (Fig. 4). Neither of the two negative control mRNAs, rpA1 and CG3083, were significantly enriched in WISP IP. This suggests that the ten WISP-targeted mRNAs and WISP protein are co-present in the same RNPs and they are likely to be directly regulated by WISP.
Figure 4. WISP forms a complex with candidate target mRNAs.
Embryos collected at 0- to 1-hr post deposition from wild type flies were fixed and cross-linked in formaldehyde for RNA immunoprecipitation. Cytoplasmic extracts from these embryos were incubated with an immobilized polyclonal anti-WISP antibody or pre-immune serum (negative control). mRNAs enriched in the IPs were isolated and used to synthesize cDNA. RT-PCR was performed to test enrichment of mRNAs in the WISP IP vs. control IP. All data were normalized to RpL32. The average of two biological replicates and one technical replicate are shown. Error bars indicate standard deviation.
Discussion
Our study demonstrates that cytoplasmic polyadenylation is a powerful mechanism for facilitating cellular change. Post-transcriptional control through poly(A) tail elongation has been observed previously on several maternal mRNAs in the cytoplasm of Drosophila oocytes and early embryos. Importantly, experimental evidence demonstrates that this polyadenylation is necessary for protein production: the test-case mRNAs that fail to get polyadenylated in wisp null mutants also fail to get translated in these mutants (Benoit et al., 2008; Cui et al. 2008). Previous studies of cytoplasmic polyadenylation in oocytes and embryos had tested a very limited number of candidate genes encoding transcripts that were expected to be WISP-regulated at these stage (e.g. cell cycle and patterning genes). In the study described here, we show that rather than being limited to a small group of specific maternal mRNAs, cytoplasmic polyadenylation is a major regulatory mechanism widely used on many maternal mRNAs at two distinct developmental stages in Drosophila. These mRNAs fail to elongate their poly(A) tails when the GLD-2 class cytoplasmic poly(A) polymerase WISP is not functional, suggesting these maternal mRNAs are putative targets of the cytoplasmic polyadenylation machinery during normal development.
During oogenesis, large amounts of gene products are packed into the developing oocyte, where they must remain quiescent until activation of embryonic development. However, there are changes that occur in the developing oocyte during late oogenesis that may require the activation of some of these gene products for completion of egg development. Prior to oocyte maturation in Drosophila, the oocyte arrests in prophase I of meiosis, and can remain arrested for several days. At maturation during late oogenesis, this arrest is relieved and the oocyte progresses to metaphase I, where it will arrest once more until the mature oocyte is ovulated. It has been shown that oocyte maturation involves the polyadenylation and translational activation of some transcripts involved in cell cycle control: PAN GU kinase activates both polyadenylation and translation of cyclin A mRNA, which promotes cell cycle progression into prometaphase (Vardy et al., 2009). Poly(A) tail elongation of cyclin B mRNA also occurs during oocyte maturation. About 100 to 120 adenosine residues are added to its mRNA (Benoit et al., 2005; Vardy and Orr-Weaver, 2007). This polyadenylation is mediated by the Drosophila homologs of known cytoplasmic polyadenylation machinery including WISP, and is believed to activate the translation of cyclin B for the exit from prophase I arrest. In the current study, we show that transcripts from over 2000 genes are likely regulated in a similar manner by WISP during late oogenesis. A substantial subset of these genes is involved in cell cycle control and chromosome organization, consistent with the cellular events that take place during oocyte maturation. Our validation studies confirm that most of the targets identified in our microarray, such as separase and CDC6, are indeed polyadenylated in a WISP-dependent manner during oogenesis. Interestingly, heat shock factor and the translation initiation factor eIF4G were also identified and confirmed as targets of WISP-dependent polyadenylation, indicating that activation of the more general regulators of cellular activity may also be critical while major cellular changes like maturation are underway.
After maturation, the Drosophila oocyte remains arrested at metaphase I of meiosis until the egg is ovulated. Ovulation induces a set of events collectively known as egg activation (Heifetz et al., 2001), which includes meiotic resumption, changes in stored proteins’ phosphorylation states, and transcript degradation or translation. These events prepare an oocyte to begin embryogenesis. Using a candidate-based approach, previous studies have found that transcripts of several genes involved in embryonic patterning are regulated by cytoplasmic polyadenylation, including bicoid, torso, Toll, and hunchback (Salles et al., 1994; Wreden et al., 1997). Our data suggest that over 4000 maternally-supplied transcripts undergo WISP-dependent polyadenylation in the cytoplasm of activating eggs, and translation of these transcripts likely facilitates the varied events of egg activation. In addition to embryonic patterning genes, mRNAs encoding cell cycle components such as grapes (Chk1 homolog), string (CDC25 homolog) and Bj1 (Rcc1 homolog) also undergo poly(A) tail extension during this period, and likely participate in meiotic resumption or subsequent embryonic mitoses. A striking finding from our early embryo data is the prevalence of targets involved in signal transduction. In many species, egg activation is triggered by a rise in intracellular calcium, often induced by factors in the fertilizing sperm. This calcium rise sets off a signaling cascade that ultimately results in the events of egg activation. In Drosophila, fertilization is not necessary for egg activation (Doane, 1960; Heifetz et al., 2001), though calcium signaling is still required for most events including meiotic resumption (Horner and Wolfner, 2008). It is plausible that cell signaling factors are translationally activated by WISP in order to carry out downstream signaling events. Our GO analysis identified enrichment of genes with functions relating to ion binding and phosphorylation, which may indicate changes in enzymatic function leading to signal transduction. In our validation tests, we positively identified the kinases Dsor1 (MEK) and Mekk1 as well as the phosphatase Pp1α-96A as WISP targets at egg activation. Activation of these classes of proteins could be responsible for signaling events at egg activation, as well as the wide range of changes in proteins’ phosphorylation states following egg activation (Krauchunas et al., 2012).
Our microarray data also indicate that transcripts of a nuclear poly(A) polymerase, hiiragi (hrg), are also polyadenylated by WISP during egg activation (Table S1); therefore, it is possible that translation of this PAP is increased as embryogenesis initiates. The nuclear PAP encoded by hrg is required in many cell types (Murata et al., 2001). During oogenesis, it is active during early to mid-stages (until stage 9–10), in contrast to WISP, whose effects are only seen at later stages (Benoit et al., 2008). During these late oogenic stages and during egg activation and early embryogenesis, de novo transcription is silenced; thus, regulation of hrg by WISP may affect polyadenylation subsequently during embryogenesis, but is not expected to impact the early-stage poly(A) population that we examined in the present study.
We identified transcripts for 2318 genes that appeared WISP-regulated in both the oocyte and the embryo. This may indicate that these transcripts undergo two rounds of polyadenylation: once at oocyte maturation, and again at egg activation. One of the transcripts we found in this category encodes Cyclin B. Previous studies showed that the poly(A) tail of this mRNA is lengthened at both stages: cyclin B mRNA was reported to have a short poly(A) tail throughout early oogenesis, but upon oocyte maturation the poly(A) tail lengthens to approximately 100 nucleotides and further lengthens to approximately 150 nucleotides at egg activation (Vardy and Orr-Weaver, 2007). The mRNA encoding Cyclin A, which was also reported to show poly(A) tail extension at both oocyte maturation and egg activation (Vardy et al., 2009), also appeared in both our WISP-dependent oocyte and embryo groups. It is not possible to use our microarray data to definitively determine which transcripts undergo two distinct rounds of polyadenylation at these two stages. However, DAVID analysis shows that many of the transcripts that we detect among WISP-regulated RNAs at both the oocyte and early embryo stages fall into cell cyclerelated categories, like cyclins A and B. We speculate that such cell cycle regulators might require WISP regulation at both times because their action is critical at both those times: earlier to promote proper maintenance of meiotic events, and later for early-embryo cell cycles. Further testing through direct poly(A) tail measurement of individual cell cycle regulators found in both WISP-dependent datasets will establish whether the cell cycle regulators identified in our studies show poly(A) tail extension at both stages, analogous to what is seen for the mRNAs encoding Cyclins A and B. In addition it will be interesting to test whether action of WISP at either or both stages promotes the ability of these target RNAs to be incorporated into polysomes.
Although specific sequence elements that mark a transcript for poly(A) tail adjustments have been well studied in vertebrates (reviewed in Belloc et al., 2008) less is known about how cytoplasmic polyadenylation is specified in Drosophila. A previous in vitro study indicated that canonical sequence elements for nuclear polyadenylation that have been identified in vertebrates, CPE (cytoplasmic polyadenylation element) and hexamer AAUAAA, are functional in the Drosophila system, but may not be necessary for cytoplasmic polyadenylation (Coll et al., 2010). Instead, some Drosophila transcripts may be regulated in a through a noncanonical polyadenylation mechanism involving a different signal sequence in the 3’UTR (Coll et al., 2010). We attempted to identify consensus sequences shared among the 3’UTRs of the candidate genes identified in our microarrays but were unable to identify a compelling motif (Figure S3). Thus, the sequences required for WISP-dependent cytoplasmic polyadenylation are not known. However, unlike canonical nuclear PAPs, Gld-2 family members lack an RNA binding domain and must form a heterodimer with a binding partner in order to polyadenylate a particular transcript (Wang et al., 2002). It is possible that WISP has multiple binding partners during oocyte maturation and egg activation, and each partner may require a different sequence element. Multiple binding partners may thus prevent us from identifying a single consensus sequence among the WISP targets.
Our microarray study has revealed that cytoplasmic polyadenylation can be a major regulator of cellular change. The regulation of oogenesis and egg activation by a GLD-2-dependent mechanism is not restricted to Drosophila, nor is the phenomenon of cytoplasmic polyadenylation in general unique to the female gametes. However, the number of WISP targets identified in our microarray—well into the thousands at two different developmental periods—is greater than any group of GLD-2 targets identified to date. The cytoplasmic polyadenylation machinery has been well dissected using Xenopus oocytes, where xGLD2 has been studied during oocyte maturation (Barnard et al., 2004). Here, similar to the case in Drosophila, xGLD2 is known to elongate the poly(A) tails of several mRNAs, such as those encoding Cyclin B and Mos, thus activating their translation (Barkoff et al., 2000; Stebbins-Boaz et al., 1996). Studies of the mouse GLD-2 homolog have also been performed. While mouse GLD-2 is present in the female germline during meiosis, knockout of mouse GLD-2 has no effect on egg maturation, suggesting that there may be a functionally redundant protein at play if cytoplasmic polyadenylation is a major regulator at this time (Nakanishi et al., 2006). As in Drosophila, C. elegans GLD-2 is involved in meiotic progression and embryogenesis (Kadyk and Kimble, 1998; Wang et al., 2002). Interestingly, C. elegans GLD-2 targets a different set of transcripts depending on the RNA-binding protein with which it is partnered (Kim et al., 2009; Kim et al., 2010). RIP-chip studies indicate that the C. elegans GLD-2 homolog has approximately 550 targets in whole adults, but the enzyme acts only on 335 of these when bound to the binding partner RNP-8 (Kim et al., 2010). Previous studies have identified the GLD-3 homolog Bic-C as a WISP binding partner in the Drosophila female germline (Benoit et al., 2008; Cui et al., 2008). Given a different binding partner, or in a different tissue, a GLD-2 enzyme may activate distinctive sets of target transcripts beyond those identified by our microarray study.
GLD-2s and other families of cytoplasmic PAPs also act outside of the female germline. The Drosophila genome encodes a second GLD-2 homolog, a paralog of wispy, that acts during post-meiotic spermatogenesis (Sartain et al., 2011). The same paralog has a second function in the brain, where it is required for long-term memory storage (Kwak et al., 2008), and there are other examples of vertebrate cytoplasmic PAPs that are required for gene expression at axon synapses (Rouhana et al., 2005). Finally, non-GLD-2 PAPs have also been identified in the mouse male germline (Kashiwabara et al., 2000). These findings indicate that the cytoplasmic polyadenylation system is a widely used mechanism for generating a quick protein production response in cell types that must undergo rapid changes, such as a firing neuron or an activating oocyte. Our findings that WISP regulates thousands of genes in this way demonstrates that the effects of cytoplasmic polyadenylation in a single cell can be very large-scale; thus, control of cytosolic mRNAs through cytoplasmic polyadenylation is a crucial mechanism for allowing coordinated, synchronous cellular change.
Supplementary Material
Highlights.
We find widespread change in mRNA polyadenylation during Drosophila egg activation.
Surprisingly, the change affects most maternally-loaded mRNAs (but not all GO classes).
The GLD2 cytoplasmic poly(A) polymerase “Wispy” causes the poly(A) elongation.
The candidate regulated-mRNAs that we tested co-IP with Wispy.
Wispy also regulates polyadenylation of many mRNAs in late-stage oocytes.
Acknowledgements
We thank A. Clark for helpful discussion and J. Sitnik, J. Liu and M. Goldberg for comments on this manuscript. We appreciate support of this work from NIH grant R01-GM044659 (to MFW). For part of this work, CVS was supported by NIH Training Grant T32-GM07617.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barkoff AF, Dickson KS, Gray NK, Wickens M. Translational control of cyclin B1 mRNA during meiotic maturation: coordinated repression and cytoplasmic polyadenylation. Dev Biol. 2000;220:97–109. doi: 10.1006/dbio.2000.9613. [DOI] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Belloc E, Pique M, Mendez R. Sequential waves of polyadenylation and deadenylation define a translation circuit that drives meiotic progression. Biochem Soc Trans. 2008;36:665–670. doi: 10.1042/BST0360665. [DOI] [PubMed] [Google Scholar]
- Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev Cell. 2005;9:511–522. doi: 10.1016/j.devcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development. 2008;135:1969–1679. doi: 10.1242/dev.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt ML, Kim AM, O'Halloran TV, Woodruff TK. Zinc requirement during meiosis I-meiosis II transition in mouse oocytes is independent of the MOS-MAPK pathway. Biol Reprod. 2011;84:526–536. doi: 10.1095/biolreprod.110.086488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnetti S, Ephrussi A. Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development. 2003;130:835–843. doi: 10.1242/dev.00309. [DOI] [PubMed] [Google Scholar]
- Chang JS, Tan L, Schedl P. The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev Biol. 1999;215:91–106. doi: 10.1006/dbio.1999.9444. [DOI] [PubMed] [Google Scholar]
- Coll O, Villalba A, Bussotti G, Notredame C, Gebauer F. A novel, canonical mechanism of cytoplasmic polyadenylation operates in Drosophila embryogenesis. Genes Dev. 2010;24:129–134. doi: 10.1101/gad.568610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Sackton KL, Horner VL, Kumar KE, Wolfner MF. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics. 2008;178:2017–2029. doi: 10.1534/genetics.107.084558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Doane WW. Completion of meiosis in uninseminated eggs of Drosophila melanogaster. Science. 1960;132:677–678. doi: 10.1126/science.132.3428.677. [DOI] [PubMed] [Google Scholar]
- Heifetz Y, Yu J, Wolfner MF. Ovulation triggers activation of Drosophila oocytes. Developmental biology. 2001;234:416–424. doi: 10.1006/dbio.2001.0246. [DOI] [PubMed] [Google Scholar]
- Horner VL, Wolfner MF. Mechanical stimulation by osmotic and hydrostatic pressure activates Drosophila oocytes in vitro in a calcium-dependent manner. Dev Biol. 2008;316:100–109. doi: 10.1016/j.ydbio.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Lee E, Kwan KM, Fenger DD, Orr-Weaver TL. The Drosophila MOS ortholog is not essential for meiosis. Curr Biol. 2004;14:75–80. doi: 10.1016/j.cub.2003.12.031. [DOI] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3'UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Kashiwabara S, Zhuang T, Yamagata K, Noguchi J, Fukamizu A, Baba T. Identification of a novel isoform of poly(A) polymerase, TPAP, specifically present in the cytoplasm of spermatogenic cells. Dev Biol. 2000;228:106–115. doi: 10.1006/dbio.2000.9894. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Kim AM, Bernhardt ML, Kong BY, Ahn RW, Vogt S, Woodruff TK, O'Halloran TV. Zinc sparks are triggered by fertilization and facilitate cell cycle resumption in mammalian eggs. ACS Chem Biol. 6:716–723. doi: 10.1021/cb200084y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kim KW, Nykamp K, Suh N, Bachorik JL, Wang L, Kimble J. Antagonism between GLD-2 binding partners controls gamete sex. Dev Cell. 2009;16:723–733. doi: 10.1016/j.devcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Wilson TL, Kimble J. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc Natl Acad Sci U S A. 2010;107:17445–17450. doi: 10.1073/pnas.1012611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauchunas AR, Horner VL, Wolfner MF. Protein phosphorylation changes reveal new candidates in the regulation of egg activation and early embryogenesis in D. melanogaster. Dev Biol. 2012;370:125–134. doi: 10.1016/j.ydbio.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JC, Wickens M. GLD2 poly(A) polymerase is required for long-term memory. Proc Natl Acad Sci U S A. 2008;105:14644–14649. doi: 10.1073/pnas.0803185105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci U S A. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nature reviews Molecular cell biology. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Murata T, Nagaso H, Kashiwabara S, Baba T, Okano H, Yokoyama KK. The hiiragi gene encodes a poly(A) polymerase, which controls the formation of the wing margin in Drosophila melanogaster. Dev. Biol. 2001;233(1):137–147. doi: 10.1006/dbio.2001.0205. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Kubota H, Ishibashi N, Kumagai S, Watanabe H, Yamashita M, Kashiwabara S, Miyado K, Baba T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol. 2010;12:447–456. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- Page AW, Orr-Weaver TL. Activation of the meiotic divisions in Drosophila oocytes. Dev Biol. 1997;183:195–207. doi: 10.1006/dbio.1997.8506. [DOI] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. Developmental role and regulation of cortex, a meiosis-specific anaphase-promoting complex/cyclosome activator. PLoS Genet. 2007;3:e202. doi: 10.1371/journal.pgen.0030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles FJ, Lieberfarb ME, Wreden C, Gergen JP, Strickland S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 1994;266:1996–1999. doi: 10.1126/science.7801127. [DOI] [PubMed] [Google Scholar]
- Salles FJ, Strickland S. Analysis of poly(A) tail lengths by PCR: the PAT assay. Methods Mol Biol. 1999;118:441–448. doi: 10.1385/1-59259-676-2:441. [DOI] [PubMed] [Google Scholar]
- Sartain CV, Cui J, Meisel RP, Wolfner MF. The poly(A) polymerase GLD2 is required for spermatogenesis in Drosophila melanogaster. Development. 2011;138:1619–1629. doi: 10.1242/dev.059618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Hake LE, Richter JD. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. The EMBO journal. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev Dyn. 2005;232:593–608. doi: 10.1002/dvdy.20297. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L, Orr-Weaver TL. The Drosophila PNG kinase complex regulates the translation of cyclin B. Dev Cell. 2007;12:157–166. doi: 10.1016/j.devcel.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Vardy L, Pesin JA, Orr-Weaver TL. Regulation of Cyclin A protein in meiosis and early embryogenesis. Proc Natl Acad Sci U S A. 2009;106:1838–1843. doi: 10.1073/pnas.0813237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2009;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Williams BC, Dernburg AF, Puro J, Nokkala S, Goldberg ML. The Drosophila kinesin-like protein KLP3A is required for proper behavior of male and female pronuclei at fertilization. Development. 1997;124:2365–2376. doi: 10.1242/dev.124.12.2365. [DOI] [PubMed] [Google Scholar]
- Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Kornbluth S. Across the meiotic divide - CSF activity in the post-Emi2/XErp1 era. J Cell Sci. 2008;121:3509–3514. doi: 10.1242/jcs.036855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.