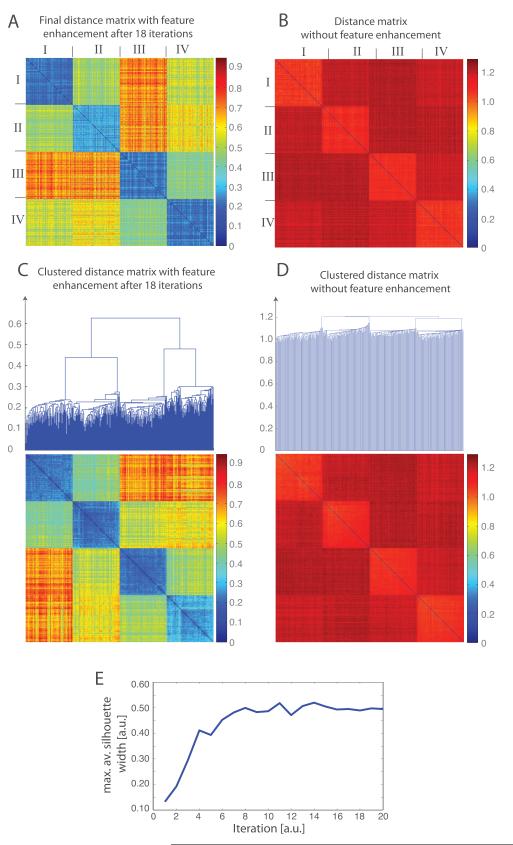
Figure 5.
Reference-free subtomogram classification for the simulated subtomograms of 4 different protein complexes as shown in Figure 4. (A) Distance matrix after 18 iterations of reference-free subtomogram classification with the feature enhancement weight . The order of subtomograms is according to the ground truth. The distance matrix is calculated using the feature enhancement weight. (B) Distance matrix calculated without feature enhancement weight after all subtomograms are aligned to their respective template density map. The order of subtomograms is according to the ground truth. (C) Clustered distance matrix after 18 iterations of the reference-free classification process using the feature-enhancement weight . Four different clusters can be clearly differentiated. (D) Clustering of the distance matrix from C. (E) Convergence of the classification process. The maximal average Silhouette width is calculated from the resulting subtomogram clusters at each iteration of the classification process. The convergence is reached when the maximal average Silhouette width reaches a plateau with increasing iterations.