Figure 2.
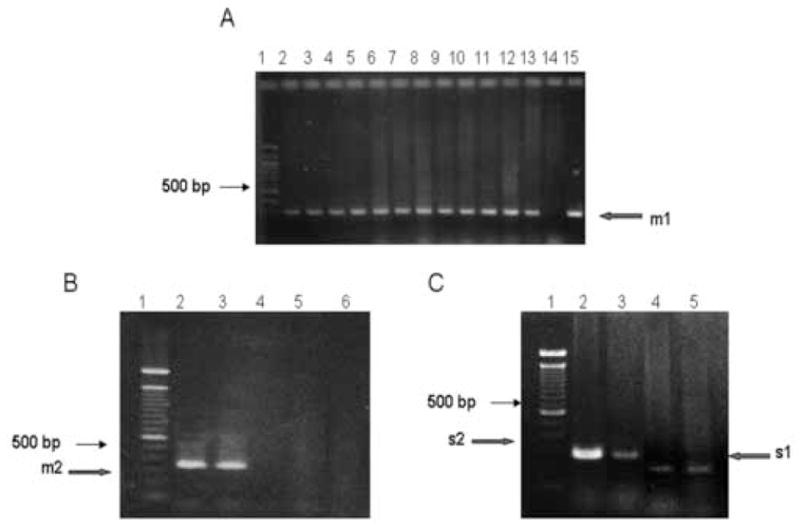
vacA amplification. A. PCR amplification of m1 allele. Lane 1: 100 bp DNA ladder standard, Lanes 2 through 13 samples obtained from patients living in IQ. Lane 14: negative control with template DNA isolated from Campylobacter jejuni. Lanes 15: positive control with chromosomal DNA isolated from strain J99. The arrow indicates the 290 bp expected PCR product. All PCR assays were repeated at least 3 times. B. PCR amplifications for m2 VacA allele. PCR fragments were separated in a 3% agarose gel. Lane 1: 100 bp ladder DNA standard, Lane 2: positive sample from a patient living in SA. Lane 3: positive control with chromosomal DNA isolated from strain 8822. Lanes 4 and 5: negative samples. Lane 6: strain 26695 as a negative control. The arrow indicates electrophoretic migration in a 3% agarose gel of the 352 bp expected fragment. C. Size differences in s1 and s2 PCR fragments derived from vacA gene from Chilean H. pylori clinical isolates. Fragments were separated in a 3% agarose gel. Lane 1: 100 bp ladder DNA standard. Lanes 2 and 3: s2 positive strains isolated from SA. Lane 4: a s1 positive strain from TE. Lane 5: a S1 positive control corresponding to H. pylori strain 26695. Arrows indicate the expected size for each fragment.
