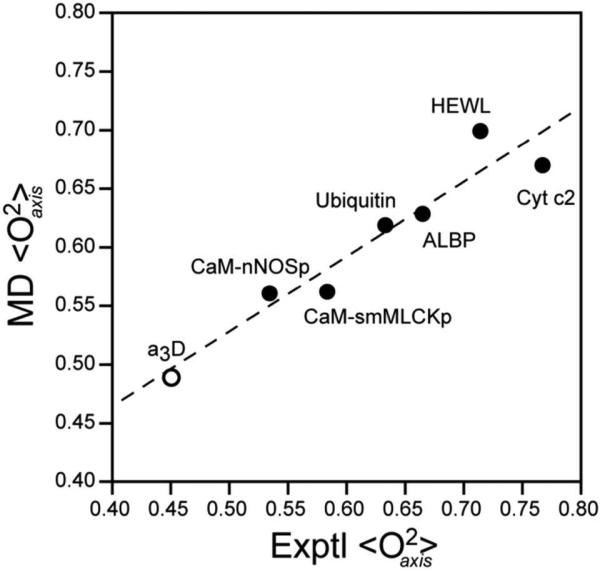
Figure 1.
Comparison of the average experimentally determined L-S squared generalized order parameters of the methyl group symmetry axis (<O2axis>) with that calculated from the molecular dynamics (MD) simulations. The experimental average includes all available data. With the exception of α3D, the molecular dynamics average includes all methyl groups. In the case of α3D, the experimentally accessible sites are compared directly with the MD average of those sites to avoid an apparent an artifact of limited experimental sampling. Linear regression yields an excellent correlation (R2 = 0.92) with slope 0.64 ± 0.09 and intercept 0.21 ± 0.05. Forcing the fitted line through an intercept of zero yields a slope 0.92 with a slightly lower R2. Individual site-to-site correlation plots for each protein are provided in the Supplementary Figure S2. The correlation coefficients and the slope values for each protein are summarized in Table 2.