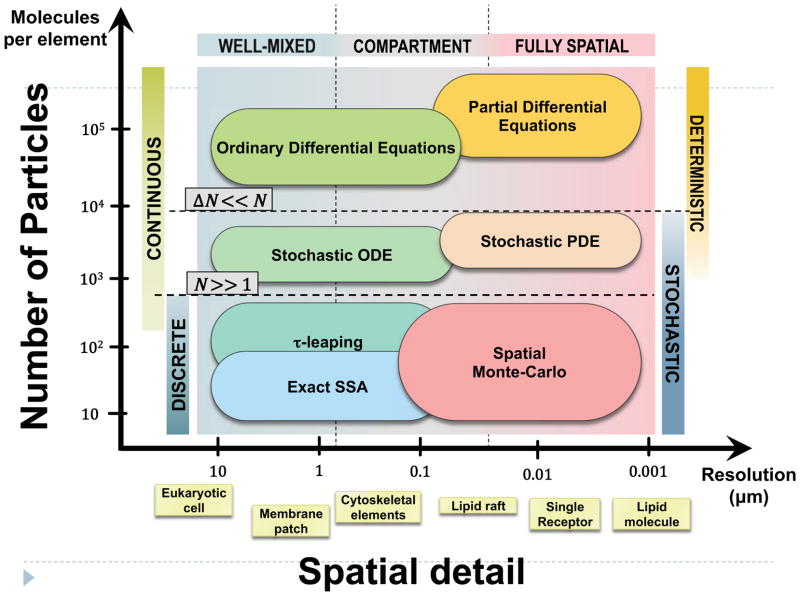
Figure 2. Classes of mathematical models for molecular processes in cells and the scales at which they are applicable to signaling processes.
A possible quantitative guide is the size of the largest element that can be treated as spatially homogeneous (horizontal axis) and the typical number of molecules of one species in the element (vertical axis). The suggested spatial resolution is determined by the size of the biological elements of interest and current computational capabilities. Spatially resolved models are resource-intensive, and are therefore generally applied to small subsystems. Cell-level models of large signaling networks are typically well mixed; spatial Monte Carlo studies rarely scale beyond a few hundred nanometers. A promising approach for multi-scale applications is a combination of compartment-based models at the large scales and fully spatial simulations focused on a few important processes within small structural elements of the membrane. Temporal fluctuations arise largely from the discrete and stochastic nature of the underlying molecular processes. The relative magnitude of temporal fluctuations (ΔN) decreases as the number of particles increases. The discrete nature of the particle number can thus be ignored when N is significantly greater than 1. That is, deviations from the expected average behavior can be neglected when the expected magnitude of the fluctuations is small compared to N.