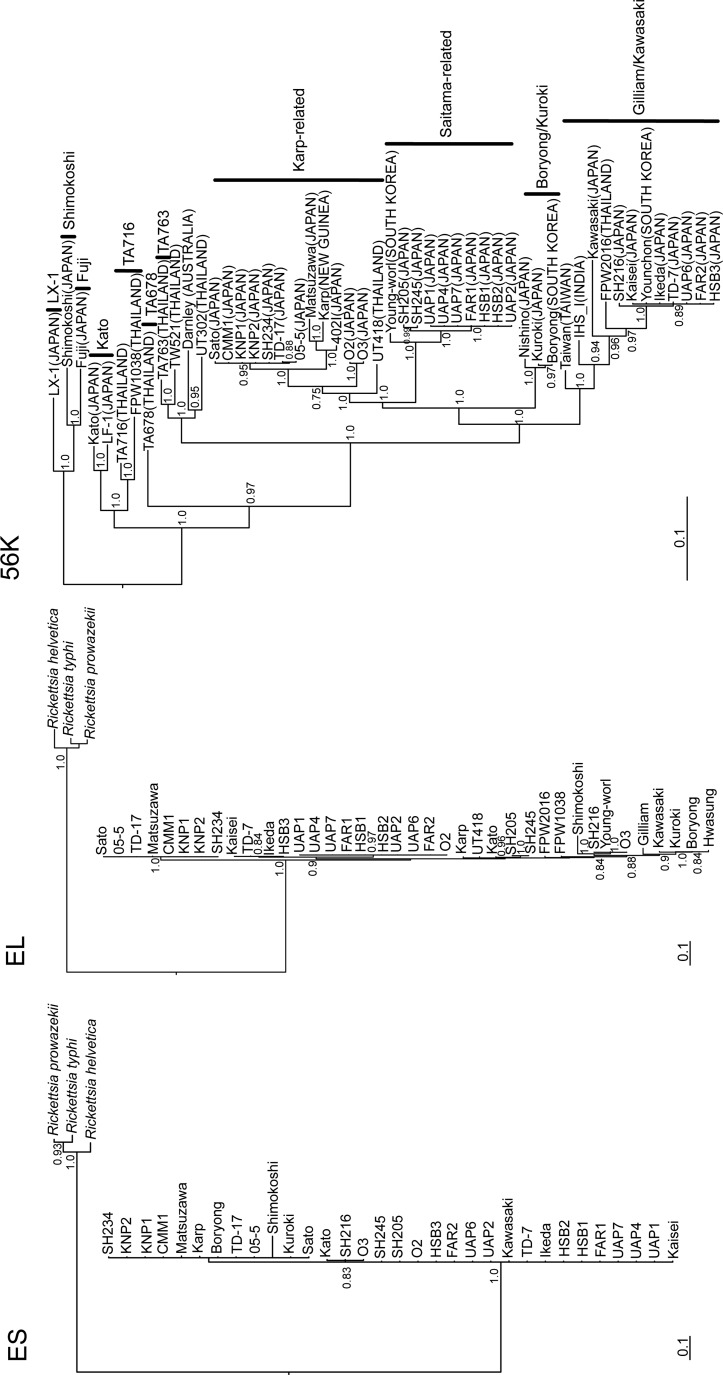
FIG. 1.
Phylogenetic trees generated using the Bayesian Markov chain Monte Carlo estimation methods (MCMC) with the GTR+I+Γ model of evolution, which was estimated from the data based on the alignment of the coding regions of the 271- to 285-nucleotide groES (ES), 1651- to 1668-nucleotide groEL (EL), and 420- to 1481-nucleotide 56-kD type-specific antigen (TSA) (56K) genes of the following Orientia tsutsugamushi strains: Kaisei (56-kD TSA, JX235719; groESL, JX188391) from patient, Sato (56-kD TSA, JX235718; groESL, JX188392) from patient, Kato (56-kD TSA, M63382; groESL, JX188393) from patient, Kuroki (56-kD TSA, M63380; groESL, JX188394) from patient, Matsuzawa (56-kD TSA, AF173043; groESL, KC688324) from patient, Shimokoshi (56-kD TSA, M63381; groESL, JX188395) from patient, UAP1 (56-kD TSA, AF302991; groESL, JX188396) from Apodemus speciosus, UAP2 (56-kD TSA, AF302992; groESL, KC688329) from A. speciosus, UAP4 (56-kD TSA, AF302993; groESL, JX188397) from A. speciosus, USP6 (56-kD TSA, AF302994; groESL, KC688325) from A. speciosus, UAP7 (56-kD TSA, AF302995; groESL, JX188398) from A. speciosus, FAR1 (56-kD TSA, AF302989; groESL, JX188399) from A. speciosus, FAR2 (56-kD TSA, AF302990; groESL, KC688326) from A. speciosus, HSB1 (56-kD TSA, AF302983; groESL, JX188400) from A. speciosus, HSB2 (56-kD TSA, AF302984; groESL, JX188401) from A. speciosus, HSB3 (56-kD TSA, AF302985; groESL, KC688327) from A. speciosus, CMM1 (56-kD TSA, AF302986; groESL, KC688328) from A. speciosus, KNP1 (56-kD TSA, AF302987; groESL, KC688320), KNP2 (56-kD TSA, AF302988; groESL, KC688321), O2 (56-kD TSA, KC688322; groESL, KC688330) from A. speciosus, O3 (56-kD TSA, KC688323; groESL, KC688331) from Apodemus argenteus, SH205 (56-kD TSA, KC693730; groESL, KC688332) from A. speciosus, SH234 (56-kD TSA, KC693731; groESL, KC688333) from A. speciosus, SH245 (56-kD TSA, KC693732; groESL, KC688334) from A. speciosus, TD-7 (56-kD TSA, JX188388; groESL, JX235722) from patient, TD-17 (56-kD TSA, JX188387; groESL, JX235720) from patient, 05-5 (56-kD TSA, JX188390; groESL, JX235721) from rodent, and SH216 (56-kD TSA, JX188389; groESL, JX188402) from Mus musculus. The phylogenetic positions of these strains are also shown relative to the following representative O. tsutsugamushi strains: Gilliam (groESL, AY191585) from patient, Taiwan (56-kD TSA, DQ485289) from patient, Ikeda (56-kD TSA, AF173033; groESL, NC010793) from patient, Younchon (56-kD TSA, U19903) from patient, Karp (56-kD TSA, M33004; groESL, M31887) from patient, Kawasaki (56-kD TSA, M63383; groESL, AY191587) from patient, Boryong (56-kD TSA, AM494475; groESL, NC009488) from patient, Nishino (56-kD TSA, AF173048) from patient, Young-worl (56-kD TSA, AF43141; groESL, AY191588) from patient, 402I (56-kD TSA, AF173047) from patient, Darnley (56-kD TSA, AY860955) from patient, FPW1038 (56-kD TSA, EF213087; groESL, EF551288) from patient, FPW2016 (56-kD TSA, EF213085; groESL, EF551289) from patient, Fuji (56-kD TSA, AF210834) from Leptotrombidium fuji, IHS I (56-kD TSA, DQ530440) from patient, Hwasung (groESL, QY191589) from patient, LF-1 (56-kD TSA, AF173050) from Leptotrombidium fletcheri, LX-1 (56-kD TSA, AF173042) from Leptotrombidium mite, TA678 (56-kD TSA, U19904) from Rattus rattus, TA716 (56-kD TSA, U19905) from Menetes berdmorei, TSA763 (56-kD TSA, RTU80636) from Rattus rajah, TW52-1 (56-kD TSA, AY222630) from Rattus norvegicus, UT302 (56-kD TSA, EF213095) from patient, and UT418 (56-kD TSA, EF213087; groESL, EF551310) from patient, with Rickettsia helvetica (DQ442911), Rickettsia prowazekii (Y15783), and Rickettsia typhi (AF462073) as outgroups. Genotypes based on the 56-kD TSA gene are shown on the right of 56K. The numbers at each node are the posterior node probabilities (>0.70) based on 150,000 trees. Two replicate MCMC runs consisting of six chains of 10 million generations were sampled every 100 generations with a burn-in of 25,000 (25%). The scale bar indicates the nucleotide substitutions per site. The country of origin is shown in parentheses.