FIG. 4.
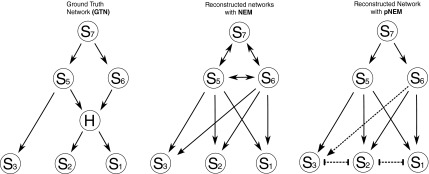
Performance comparison: The left part of the figure shows a GTN that has one hidden node. We attach a total of 350 E-genes uniformly to the S-genes and generate artificial data using a moderate noise level of 0.15 for both false negative and false positive observations. The plots in the middle and on the right side of the figure show the reconstructed network using triplet search in a standard NEM approach and pNEM, respectively, only for the observable nodes. The NEM incorrectly predicts a feedback loop–like structure. In contrast, the pNEM resolved the relation between S5 and S6 as R5, thus predicting the existence of the hidden node at that position. Also, all other predicted relations are correct, with the exception of S6 and S3, where the pNEM is undecided whether a directed relation exists (incorrect) or not (correct).
