Abstract
Tissue factor pathway inhibitor-2 (TFPI-2) is a potent inhibitor of plasmin which activates matrix metalloproteinases (MMPs) involved in degradation of the extracellular matrix. Its secretion in the tumour microenvironment makes TFPI-2 a potential inhibitor of tumour invasion and metastasis. As demonstrated in aggressive cancers, TFPI-2 is frequently down-regulated in cancer cells, but the mechanisms involved in the inhibition of tumour progression remained unclear. We showed in this study that stable TFPI-2 down-regulation in the National Cancer Institute (NCI)-H460 non-small cell lung cancer cell line using specific micro interfering micro-interfering RNA promoted tumour progression in a nude mice orthotopic model that resulted in an increase in cell invasion. Moreover, TFPI-2 down-regulation enhanced cell adhesion to collagen IV and laminin via an increase in α1 integrin on cell surface, and increased MMP expression (mainly MMP-1 and -3) contributing to cancer cell invasion through basement membrane components. This study also reveals for the first time that pulmonary fibroblasts incubated with conditioned media from TFPI-2 silencing cancer cells exhibited increased expression of MMPs, particularly MMP-1, -3 and -7, that are likely involved in lung cancer cell invasion through the surrounding stromal tissue, thus enhancing formation of metastases.
Keywords: tissue factor pathway inhibitor-2, micro interfering RNA (miRNA), non-small cell lung cancer, tumour growth, cell invasion, cell adhesion, integrins, matrix metalloproteinases
Introduction
Tumour progression is a complex multistep process that depends on an evolving crosstalk between cancer cells and the surrounding stromal tissue. The microenvironment is now recognized as having a pivotal role in promoting cancer initiation, progression and dissemination to form metastases [1]. The invasion process involves extracellular matrix (ECM)-degrading proteases, particularly matrix metalloproteinases (MMPs), that have been shown to be highly expressed and activated in the tumour microenvironment [2], especially in highly aggressive malignant tumours [3, 4]. Activated fibroblasts, the major cell component of the microenvironment, actively contribute to tumour invasiveness by secreting a consistent amount of MMPs at the tumour–stroma interface. Moreover, this MMP synthesis could be increased by the ECM metalloproteinase inducer (EMMPRIN) expressed by cancer cells. Activation of zymo-gens (pro-MMPs) in the extracellular environment needs proteolytic cleavage of the aminoterminal prodomain [5] that depends on ser-ine proteases, such as trypsin and plasmin, and also involves activated MMPs or membrane-anchored matrix metalloproteinases (MT-MMPs). Apart from tissue inhibitors of metalloproteinases (TIMPs) that are specific regulators of MMP activity, TFPI-2 (tissue factor pathway inhibitor-2), an inhibitor of serine proteases – particularly plasmin – could also regulate the activation of MMPs [6, 7], thus regulating ECM degradation and tumour cell invasion.
TFPI-2 is a 32 kD Kunitz-type serine proteinase inhibitor secreted into the ECM by a wide variety of human cells including endothelial cells, monocytes, fibroblasts, epithelial cells, smooth muscle cells, syncytiotrophoblast cells [8–11] and also several human tumour cells [12–15]. Interestingly, TFPI-2, now considered to be a candidate tumour suppressor gene, has been shown to be down-regulated in particularly aggressive tumours [11, 13, 16] in association with epigenetic changes. Hypermethylation of CpG islands of TFPI-2 promoter and histone deacetylation have frequently been correlated with transcriptional silencing of the TFPI-2 gene in cancer cells [16–21]. In agreement with these studies, we have demonstrated that decreased TFPI-2 gene expression and hypermethylation are frequently associated with advanced stages of non-small cell lung cancer, particularly with lymph node invasion [15]. The consequences of TFPI-2 down-regulation on MMP expression by both tumoural and stromal cells remain unclear and could have a key role in controlling tumour invasion by modifying the balance between ECM-degrading proteases and their inhibitors into the microenvironment.
We therefore investigated the impact of stable TFPI-2 inactivation in NCI-H460 non-small lung cancer cells on their behaviour toward lung fibroblast cells. We applied the promising new micro-interfering RNA approach (miRNA) to trigger sequence-specific TFPI-2 RNA degradation and gene silencing. We studied the effects of TFPI-2 inactivation on tumour growth in a nude mice orthotopic model and then invasiveness, proliferation and adhesion properties of cancer cells to ECM proteins and their MMP expression pattern. We also evaluated whether TFPI-2 inactivation in cancer cells might be responsible for regulation of MMP synthesis by pulmonary fibroblasts.
Materials and methods
Cell cultures
The human non-small cell lung cancer cell line NCI-H460 was obtained from the American Type Culture Collection (LGC Promochem, Molsheim, France). Cells were grown in Roswell Park Institute Medium 1640 medium (Invitrogen, Cergy-Pontoise, France) supplemented with 2 mM L-glutamine, 25 mM sodium bicarbonate, 2 mM glucose, 10 mM 4-(2-hydroxyethyl)-1-piperazi-neethanesulfonic acid (HEPES), 1 mM sodium pyruvate, 100 μg/ml streptomycin, 100 U/ml penicillin and 10% endotoxin-free heat inactivated foetal calf serum (FCS, ATGC Biotechnologie, Noisy le Grand, France). The human fibroblast cell line CCD19-Lu (LGC Promochem) derived from adult normal lung tissue was grown in MEM/Earle's/Glutamax medium supplemented with non-essential amino acids, 25 mM sodium bicarbonate, 1 mM sodium pyru-vate, 100 μg/ml streptomycin, 100 U/ml penicillin and 10% FCS. All cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C.
Construction of pre-micro-interfering RNA targeting TFPI-2
Two different sequences of pre-miRNA targeting the human TFPI-2 transcripts [22] were designed using Invitrogen's RNAi design algorithm and BLAST to avoid off-target gene silencing. Pre-miRNA-1 sequence targets the nucleotides 858 to 878 of the 3′UTR region. The sequence of pre-miRNA-2 was designed according to the TFPI-2 siRNA we used in a previous study [23] and targets the nucleotides 581 to 601 of the TFPI-2 K3 domain.
The pcDNA 6.2-GW/EmGFP-miR plasmid (Invitrogen) with blasticidin resistance gene and expressing the EnGFP (emerald green fluorescent protein) was used for the synthesis of pre-miRNA. This pre-miRNA is based on the murine miR-155 sequence [24] and the stem loop structure was optimized by the supplier to obtain a high knockdown rate. Pre-miRNA is then processed by endogenous Dicer enzyme into a 22 nucleotide mature miRNA using cellular machinery. The sequences used were: miRNA-1, top strand 5′-tgctgatgatttgtttcctcatgctggttttggccactgactgaccagcatgaaaacaaatcat-3′; bottom strand 5′-cctgatgatttgttttcatgctggtcagtcagtggccaaaaccagcatgaggaaacaaatcatc-3′; miRNA-2, top strand 5′-tgctgaataatagcgagtcacattgggttttggccactgactgacccaatgtgtcgctattatt-3′; bottom strand 5′-cct-gaataatagcgacacattgggtcagtcagtggccaaaacccaatgtgactcgctattattc-3′. Sequences were annealed and ligated using 1 U/μl T4 DNA ligase (Invitrogen). A control plasmid vector expressing miRNA not directed against a known mammalian gene was used as a negative control (miRNA-Neg). The recombinant plasmids were recovered by PCR using the forward sequencing primer (5′-ggcatggacgagctgtacaa-3′) and the reverse sequencing primer (5′-ctctagatcaaccactttgt-3′) (Invitrogen). The PCR product sequences which contained the miRNA inserts were sequenced (Perkin Elmer AbiPrism™ 377 DNA sequencer).
Stable transfection with plasmids encoding TFPI-2 pre-miRNA
Confluent NCI-H460 cells were washed with Ca2+ and Mg2+-free Hank's balanced solution and harvested using 0.05% trypsin-0.02% ethylenediaminetetraacetic acid (EDTA). Cell viability was determined by Trypan blue dye exclusion test and ranged between 90% and 95%. For miRNA transfection, 105 cells were seeded in 24-well plates for 24 hrs in RPMI-1640 10% FCS without antibiotics. Purified pcDNA 6.2GW/EmGFP-miR expression vectors (0.8 μg) containing either the TFPI-2 pre-miRNA insert (pcDNA-TFPI-2 pre-miRNA-1, pcDNA-TFPI-2 pre-miRNA-2) or a negative-control mismatch sequence (pcDNA-TFPI-2 pre-miRNA-Neg) were transfected into 75–80% confluency NCI-H460 cells with 2 μl of Lipofectamine 2000 reagent (Invitrogen). Six hours after transfection, the medium was replaced by fresh complete medium containing 10% FCS. After 24 hrs, cells were plated in 6-well plates with selection medium, containing 6 μg/ml blasticidin. Transfection efficacy was checked by fluorescence microscopy 48 hrs after transfec-tion by measuring EmGFP expression. Successfully transfected cell clones were then obtained by 3 weeks culture in the selection medium and TFPI-2 knockdown was assessed by reverse transcriptase real-time PCR and Western blotting.
Reverse transcription and real-time PCR
Total mRNA was extracted from 106 cells using the Dynabeads mRNA Direct Kit (Invitrogen) according to the manufacturer's instructions. Total mRNA was then reverse transcribed for 1 hr at 42°C in incubation buffer containing 250 μM of each deoxynucleotide triphosphate, 5 μM oligo (dT)20, 24 units RNase inhibitor, and 20 units of avian myeloblastosis virus reverse transcriptase (Roche Diagnostics, Meylan, France).
The amounts of TFPI-2, MMP-1, -2, -3, -7, -9, -13, -14 and EMMPRIN transcripts within cells were assessed by real-time PCR using the icycler iQ detection system (Bio-Rad, Ivry sur Seine, France). PCR was performed in a total reaction volume of 25 μl containing cDNA obtained from mRNA of 2 × 104 cells, 2-fold dilution of Platinum Quantitative PCR SuperMix-UDG (Invitrogen), 0.32 μM of each primer (Eurogentech, Angers, France, Table 1) and a 50,000-fold dilution of Sybr Green solution (Roche Diagnostics). To study gene expression, PCR were initiated by decontamination (50°C for 2 min.) and denaturation steps (95°C for 2 min.), followed by n cycles (Table 1) at 95°C for 20 sec. and at hybridization T°C for 40 sec. The melting curve was analysed for each sample to check PCR specificity. For each gene studied, specific standard curves were established using decreasing amounts of purified PCR products (from 107 to 5 × 101 copies). mRNA copies of the gene of interest were then normalized to 106 copies of β-actin mRNA used as gene control.
Table 1.
Oligonucleotide sequences used for real-time PCR
| Gene | Sequences (5′ 3′) | Size (bp) | Hybridization T°C | Cycles |
|---|---|---|---|---|
| TFPI-2 | Forward: AACGCCAACAATTTCTACACCT Reverse: TACTTTTCTGTGGACCCCTCAC | 109 | 67 | 35 |
| MMP-1 | Forward: CTGCTGCTGTTCTGGGGT Reverse: GCCACTATTTCTCCGCTTTTC | 147 | 65 | 40 |
| MMP-2 | Forward: GGCCCTGTCACTCCTGAGAT Reverse: CAGTCCGCCAAATGAACCGG | 105 | 67 | 34 |
| MMP-3 | Forward: ATCCCGAAGTGGAGGAAAAC Reverse: GCCTGGAGAATGTGAGTGGA | 139 | 65 | 40 |
| MMP-7 | Forward: CCGCATATTACAGTGGATCG Reverse: GCCAATCATGATGTCAGCAG | 111 | 60 | 40 |
| MMP-9 | Forward: AGACCGGTGAGCTGGATAG Reverse: GTGATGTTGTGGTGGTGCC | 121 | 69 | 45 |
| MMP-13 | Forward: AGCATGGCGACTTCTACCC Reverse: CATCAAAATGGGCATCTCCT | 96 | 65 | 40 |
| MMP-14 | Forward: CGAGGGGAGATGTTTGTCTT Reverse: TCGTAGGCAGTGTTGATGGA | 131 | 65 | 40 |
| EMMPRIN | Forward: TGCTGGTCTGCAAGTCAGAG Reverse: GCGAGGAACTCACGAAGAAC | 123 | 65 | 40 |
| β-actine | Forward: GCCCTAGACTTCGAGCAAGA Reverse: AGGAAGGAAGGCTGAAGAG | 143 | 62 | 25 |
Western blotting
Transfected cells (miRNA-1 and -2, miRNA-Neg clones and parental NCI-H460) were grown in complete medium to 70–80% confluency in 6-well plates. Cells were eliminated by trypsinization and proteins from the ECM were solubilized in TNC buffer (50 mM Tris-HCl pH 7.5, 0.15 M NaCl, 10 mM CaCl2 and 0.05% Brij 35) and then centrifuged at 15,000 × g for 5 min. Total protein concentrations of the supernatants were measured using the Lowry method (Total Protein Kit, Sigma Aldrich, Saint Quentin Fallavier, France). Proteins (3 μg) were separated on 12% SDS-PAGE and transferred onto a nitrocellulose membrane. Membranes were then saturated for 2 hrs at room temperature in TNT buffer (10 mM Tris-HCl and 150 mM NaCl pH 7.4, 0.1% Tween-20) with 5% non-fat dried milk, incubated overnight at 4°C with polyclonal rabbit anti-TFPI-2 antibody (generous gift of W. Kisiel) diluted 1/3000 in TNT buffer with 5% non-fat milk and for 1h with peroxidase-labelled anti-rabbit IgG (Sigma Aldrich) after washing with TNT buffer. Following exposure for 1 min. to the Chemiluminescence Reagent Plus (Perkin Elmer Biosystems, Courtaboeuf, France), membranes were drained, wrapped in a plastic bag and exposed to autoradiography film (Sigma Aldrich) for 10 min. in the dark.
Nude mice orthotopic model of human lung cancer
Pathogen-free male BALB/c nude mice 4 weeks old (Charles River laboratories, Lyon, France) were acclimatized for 2 weeks before starting the study in a sterile environment. All animals were handled and cared in accordance with the national and institutional guidelines. Protocols were conducted under the supervision of an authorized investigator with the approval of the institutional ethic committee where experiments are performed (CIPA, TAAM-UPS44 Orléans, France). Mice were maintained in sterilized filter-stopped cages throughout the experimentation. They were examined daily and monitored for signs of distress, decreased physical activity and weight.
Before implantation, confluent miRNA-1 and -2 and miRNA-Neg NCI-H460 cells were harvested using 0.05% trypsin and 0.02% EDTA and washed twice in FCS-free medium. Cells were then resuspended in RPMI-1640 containing 10 mM EDTA (Sigma Aldrich) added immediately before implantation. Trypan Blue dye exclusion test was used to assess cell viability >95% for implantation.
The intrabronchial tumoral cell implantation procedure was derived from that previously described [25]. Mice (nine animals for each miRNA clones) were anesthetized and the surgery area prepared with a skin disinfection using betadine swabs. Immediately before transplantation, a 99mTc-labelled tin colloid, used as tracer, was added to cell suspension containing 10 mM EDTA. A 0.5 cm ventral incision was made over the region of the trachea superior to expose the trachea that was punctured using a 23-gauge needle. Cell inoculum (7.5 × 105 tumour cells in 25 μl) was aspirated in a 1.9 Fr × 50 cm blunt-ended catheter (Beckton Dickinson, Le Pont de Claix, France) that was inserted and advanced preferentially into the right main bronchus. Position of the catheter was monitored using high resolution radiological imaging (MX-20, Faxitron X-ray Corporation, Wheeling, IL, USA). Tumour cells were slowly injected into the lung and the scintigraphic assessment of cell deposition into lung was performed (Gamma Imager, Biospace Mesures, Paris, France). The catheter was removed, the incision closed and the animals were placed on a heating pad (37°C) until fully awake. Animal reactions were observed to ensure recovery from the anaesthesia. To document tumour location and measurement, computed tomography was performed on anesthetized animals using a small animal imager (eXplore Locus, General Electric Healthcare, Velizy, France). Width (W, in axial), height (H, in mid sagittal) and length (L, in mid sagittal) were measured and the volume was calculated using the ellipsoid formula 4/3π (W/2 ×H/2 ×L/2).
Migration and invasion assays
Cell migration was assessed using a model based on the Boyden chamber (8 μm pore size, BD Biosciences). Briefly, transfected cells were harvested by trypsinisation, then washed with phosphate-buffered saline (PBS) and 2 × 105 cells were suspended in 400 μl of serum-free medium and seeded on the upper chamber of the inserts. The lower chamber of a 24-well cell culture plate was filled with 600 μl of culture medium containing 10% FCS used as chemoattractant. Plates were incubated for 48 hrs at 37°C in a humidified atmosphere containing 5% CO2. The cells remaining in the insert were removed by aspiration and wiping with cotton swabs. Migrated cells on the lower surface of the filter and adhering to the plate were detached by 0.05% trypsin-0.02% EDTA and counted in a Malassez chamber. The results were expressed as the percentage of migrating cells ± S.E.M.
To study the cell invasion through the basement membrane components, cell culture inserts were coated with a thin layer of 0.8 mg/ml Matrigel™ (BD Biosciences) according to the manufacturer's instructions.
Proliferation assay
Tumour cells (1.25 ± 104) were seeded in 24-well plates and cultured in 700 μl complete medium containing 10% FCS. After 24, 48, 72 and 96 hrs of culture, 140 μl of MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2H-tetrazolium, inner salt) and an electron coupling agent, phenazine ethosulphate (CellTiter 96® AQueousOne Solution, Promega, Charbonnières les Bains, France) were added. Cells were incubated for 1 hr at 37°C in a humidified atmosphere containing 5% CO2 and absorbance was then measured on an ELISA plate reader (Thermomax Molecular Devices, St Grégoire, France) at a wavelength of 490 nm.
Extracellular matrix protein attachment assay
Adhesion of stably pre-miRNA-transfected NCI-H460 cells to laminin, vitronectin, fibronectin and collagen I and IV was studied using the ‘CytoMatrix SCREEN Kit’ (AbCys, Paris, France) according to the manufacturer's instructions. Cells were serum-starved overnight, detached with 0.05% trypsin-0.02% EDTA and allowed to express novel integrins in the shaking incubator at 37°C in 5% CO2 for 2 hrs in complete medium containing 1% bovine serum albumin (BSA). After counting cells in the presence of Trypan blue using a Malassez chamber, 5 × 104 viable cells in 100 μl complete medium with 1% BSA were plated in triplicate on ECM protein-coated wells, or on wells coated with BSA used as control, for 2 hrs at 37°C in a humidified CO2 atmosphere. Cells were then rinsed three times with PBS containing Ca2+ and Mg2+ and adhering cells were fixed and stained in 100 μl of 0.2% crystal violet in 10% ethanol over 5 min. of gentle shaking at room temperature. Relative attachment of cells was determined using absorbance readings at 570 nm in a microplate reader (Thermomax Molecular Devices). Readings measured with BSA for each cell type were subtracted from ECM protein readings.
Identification of cell surface Integrins
Identification of cell surface integrins was performed by using the fluorimetric Alpha/Beta Integrin-Mediated Cell Adhesion Assay Combo Kit (Chemicon Millipore, Saint Quentin en Yvelines, France). Briefly, 3 × 106 tumour cells (miRNA-Neg, miRNA-1b or miRNA-2b NCI-H460 clones) were seeded in medium complemented with 1% BSA and incubated overnight at 37°C in 5% CO2. Cells were then detached with 0.05% to 0.02% EDTA and washed twice in Hank's Buffered Salt Solution (HBSS) (without Ca2+ and Mg2+). After centrifugation at 800 × g, the pellet was resuspended in 2 ml medium supplemented with 1% BSA and the cells were incubated rocking for 2 hrs at 37°C in 5% CO2. The cells were then spun down and the pellet was resuspended in 2 ml Assay Buffer. 100 μl of the cell suspension in Assay Buffer were distributed in a microtitre plate coated with anti-human Integrin mouse monoclonal antibodies and incubated for 2 hrs at 37°C in 5% CO2. The unbound cells were washed away and adhering cells were lysed and detected with the patental CyQuant GR dye (Molecular Probes, Invitrogen, Carlsbad, CA, USA), which binds to nucleic acids, by reading fluorescence with a Wallac 1420 Victor2 apparatus (excitation: 485 nm; emission: 535 nm). Results are expressed in relative fluorescence units.
Identification of regulated signal transduction pathways
The measurement of 10 signal transduction pathways in the tumour cell clones was performed with the Cignal Finder™ Cancer 10-Pathway Reporter Array (SA Biosciences, TEBU, Le Perry en Yvelines, France). Briefly, 5 × 104 NCI-H460 miRNA-Neg, miRNA-1b or miRNA-2b cells were reverse-transfected with SureFECT reagent directly in a 96-well plate containing a mixture of a pathway-focused transcription factor-responsive. Each of the 10-pathway reporter assays contains an inducible transcription factor responsive firefly luciferase reporter and constitutively expressing Renilla construct (20:1). The cells were incubated for 16 hrs in the trans-fection medium and then cultured 24 hrs in complete medium to allow the transcription factor activity on the reporters. The reporter activity is measured using the Dual Glo® Luciferase Assay kit from Promega with a Wallac 1420 Victor2 apparatus (10 sec. reading). Results are expressed in Relative Luciferase Activities (ratio firefly/Renilla).
Preparation of tumour cell-conditioned medium
Tumour cells (miRNA-Neg, miRNA-1b or miRNA-2b NCI-H460 clones) were grown in complete medium to confluency. Cells were then washed twice with HBSS medium without Ca2+ and Mg2+ and cultured with serum-free medium supplemented with 1% Nutridoma® (Roche Diagnostics) for 24 hrs at 37°C. The conditioned medium was collected, centrifuged at 1600 × g for 10 min. and concentrated 10-fold (Amicon concentrator, Millipore, Saint Quentin en Yvelines, France). Fibroblast cells seeded overnight in 6-well plates (106 cells) in complete medium with 10% FCS were washed twice with PBS, and fresh concentrated conditioned medium equivalent to 107 tumour cells (ratio 10:1) was then applied for 24 hrs. A control condition was assayed by using non-conditioned medium complemented with 1% Nutridoma® and concentrated 10-fold. Twenty-four hours later, fibroblast cells were lysed, total mRNA was extracted and the amounts of MMP-1, -2, -3, -7, -9, -13, -14 and EMMPRIN transcripts assessed by reverse transcription and real-time PCR. MMP-1, -3 and -7 protein expression was evaluated by immunofluorescence assay.
Immunofluorescence assay
Tumour cells or fibroblasts were seeded in 8-well chamber slides (LabTek, Dominique Dutscher, Issy les Moulineaux, France) 24 hrs prior to immuno-fluorescence staining in complete medium with 10% FCS or with fresh concentrated conditioned medium from tumour cells. Cells were then washed twice with PBS and fixed in 4% paraformaldehyde solution for 10 min. at room temperature. After two washings in PBS, slides were blocked with a fresh saturation solution (PBS/BSA 2%) for 1 hr and then incubated at room temperature with primary antibodies, i.e. polyclonal rabbit anti-TFPI-2 antibody (1/3 000), rabbit monoclonal anti-MMP-1 (RB-9225-P, 1/100) (Microm Microtech France, Francheville, France), anti-MMP-3 (MS-810-P, 1/100) or mouse monoclonal anti-MMP-7 (MS-813-P1, 1/200) (Microm Microtech France) for 1 hr. Cells were washed three times, followed by 1 hr incubation with AlexaFluor 488-labelled anti-rabbit IgG or antimouse IgG secondary antibodies at 1/1 000 (Molecular Probes, Invitrogen) at room temperature in the dark. For the negative control, cells were incubated with isotype-matched control antibody or secondary antibody only. Finally, four washings were performed with PBS. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) and cells were viewed under a fluorescence microscope (Leica Microsystems, Nanterre, France) using Nikon Lucia G v.5.0 software.
Statistical analysis
Data were expressed as means ± S.E.M., statistical analysis was carried out using Student's t-test (two tailed) and P < 0.05 indicates statistical significance.
Results
MicroRNA-mediated RNAi inhibits TFPI-2 expression in non-small lung cancer cells
NCI-H460 cells from a human non-small cell lung cancer were stably transfected with two recombinant plasmids encoding pre-miRNA, i.e. pre-miRNA-1 and pre-miRNA-2, targeting respectively a region of the 3′UTR and the K3 domain of the human TFPI-2 transcript, or with non-silencing miRNA showing no known homology to mammalian genes and used as negative control (miRNA-Neg). First, and in order to identify successful construction of recombinant plasmids, PCR assays were performed and the PCR product sequences containing those of the pre-miRNA inserts were verified by DNA sequencing. According to the computer analysis, these inserted nucleotides should specifically bind to homologous sites of TFPI-2 mRNA, and thus knockdown TFPI-2 expression in the NCI-H460 cells. Efficacy of transfection of NCI-H460 cells with the recombinant plasmids was evaluated with EmGFP fluorescence after cell culture for 3 weeks in medium with 6 μg/ml blasticidin in order to isolate stably transfected clones. For each miRNA, two cell clones (a and b) with the highest GFP expression rate were retained for the next experiments.
To examine miRNA-induced gene down-regulation, total mRNA and protein from the ECM of NCI-H460, miRNA-Neg, miRNA-1a and -1b, and miRNA-2a and -2b cells were extracted. Although the miRNA-Neg and miRNA-1a clones exhibited no interference effect, significant inhibition of TFPI-2 transcripts was achieved in the miRNA-1b, -2a and -2b cell clones as demonstrated by RT and real-time PCR, with 91%, 73% and 93.5% inhibition, respectively, compared to parental NCI-H460 cells (Fig. 1A). As shown in Fig. 1B, TFPI-2 protein level was strongly decreased in the ECM of miRNA-1b and -2b cells, and to a lesser extent in miRNA-2a cell ECM. The presence of TFPI-2 triplets was demonstrated in the ECM of untransfected NCI-H460 and in miRNA-Neg and miRNA-1a transfected cells. Two clones, miRNA-1b and -2b, with the highest TFPI-2 inhibition were thus selected for further experiments, and immunofluores-cence staining clearly showed the reduced expression of TFPI-2 in both these cell clones (Fig. 1C). In contrast, no variation in TFPI-2 expression was observed in miRNA-Neg cells compared to NCI-H460 parental cells. Therefore, miRNA-Neg was used as negative control. Transfection stability with miRNA was checked every month by measuring EmGFP fluorescence and the inhibition of TFPI-2 transcripts was monitored by real-time RT-PCR. No detectable loss of TFPI-2 inhibition was observed over a year.
Fig 1.
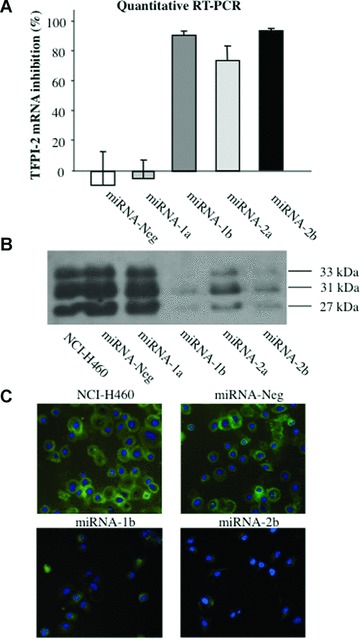
TFPI-2 silencing in lung cancer cells by micro-RNA interference. (A) TFPI-2 transcript levels were quantified using real-time RT-PCR in non-small lung cancer cells stably transfected with recombinant plasmids containing miRNA inserts that target TFPI-2 mRNA in parental NCI-H460. The copy number of TFPI-2 transcripts was first normalized to 106 copies of β-actin mRNA and the percentage of TFPI-2 mRNA inhibition in miRNA clones was compared to parental NCI-H460. Data are presented as means ± S.E.M. from four independent mRNA extractions and real-time RT-PCR performed in triplicate. (B) TFPI-2 protein expression in ECM from NCI-H460 non-small lung cancer cells and miRNA clones by immunoblotting using a rabbit polyclonal antibody against TFPI-2. (C) Immunofluorescence staining of parental NCI-H460 cells and miRNA-1b and -2b clones using a rabbit polyclonal antibody against TFPI-2 (original magnification, ×40).
Down-regulation of TFPI-2 promotes tumour progression in a nude mice orthotopic model
miRNA-Neg, miRNA-1b and -2b NCI-H460 cell suspensions containing a 99mTc-labelled tin colloid used as tracer were implanted intrabronchially with a catheter inserted into the trachea. The catheter position into the right main bronchus was checked using X-ray imaging and the punctual deposition of cells into the lung assessed by scintigraphy. Only one mouse died immediately after surgery and another one died during experimentation due to tumour progression. When cells were orthotopically implanted with 10 mM EDTA, lung tumours were obtained in 95% of mice after a 4-week period. Tumours were located in the lower and middle parts of the lung as demonstrated by tomography scanning (Fig. 2). Lung tumour volumes were calculated on axial transverse sections. The results showed an increase in tumour volumes with both miRNA-1b (P= 0.05) and -2b (P= 0.02) clones compared to miRNA-Neg cells.
Fig 2.
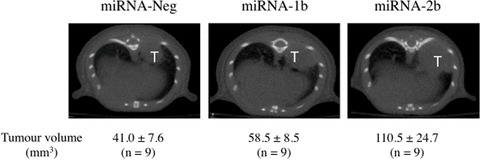
Impact of TFPI-2 silencing on tumour progression in a nude mice orthotopic model. The intrabronchial implantation of cells was performed with a 1.9 Fr × 50 cm blunt-ended catheter that was inserted and advanced into the right main bronchus. Position of the catheter was monitored using X-ray imaging. Tumour cell suspension (7.5 × 105 miRNA-Neg, miRNA-1b and -2b tumour cells in 25 μl) containing a 99mTc-labelled tin colloid and 10 mM EDTA was slowly injected into the right lobe. The scintigraphic assessment of the cell deposition into lung was then performed. Tumour progression was monitored using computed tomography scanning imaging over a 4-week period. Twenty minutes prior imaging, 100 mg/kg D-luciferin was injected i.p. to anesthetized mice. Lung tumour (T) volumes were measured on axial transverse sections. Each scanning image is representative of results obtained in nine animals per group. Tumour volume results are mean ± S.E.M. (n= 9).
TFPI-2 down-regulation promotes lung cancer cell migration and invasion without impact on cell proliferation
The migratory ability of miRNA-1b and -2b NCI-H460 clones through insert filters of 8 μm pore size was significantly increased 1.7-fold (P < 0.001) and 1.4-fold (P < 0.05) respectively), compared to those of the miRNA-Neg clone. Down-regulation of TFPI-2 was also associated with a significant increase (P > 0.001) in the invasion of miRNA-1b (2.5-fold) and of -2b (1.8-fold) cell clones through Matrigel™, which mimics basement membrane components (Fig. 3A). In both cases, the miRNA-1b cell clone exhibited greater migratory and invasion ability than miRNA-2b.
Fig 3.
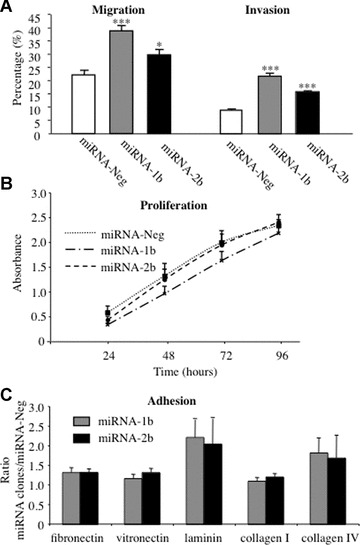
Lung cancer cell migration, invasion, proliferation and adhesion to ECM proteins. (A) Cell migration and invasion were evaluated by modified Boyden chamber assay using culture inserts (8 μm pore size) coated with a thin layer of Matrigel (0.8 mg/ml) for invasion and uncoated inserts for migration. Cells (2 × 105) were seeded in the upper chamber of the inserts and allowed to migrate for 48 hrs to the lower chamber containing culture medium with 10% FCS used as chemoattractant. Results from six experiments (means ± S.E.M.) are expressed as the percentage of migrating miRNA-1b and -2b cells compared with miRNA-Neg cells (*P < 0.05; **P < 0.001; ***P < 0.0001, Student t-test). (B) The proliferation potential of 1.25 × 104 lung cancer cells transfected with miRNA targeting TFPI-2 was evaluated at 24, 48, 72 and 96 hrs using MTS assay. Results are means (±S.E.M.) of four experiments performed in triplicate for each time-point (P-values were determined using a Student t-test). (C) Tumour cells (2 × 105) transfected with miRNA targeting TFPI-2 were allowed to adhere to fibronectin, vitronectin, laminin, collagen I and collagen IV or BSA used as control for 2 hrs and then stained with crystal violet. Absorbance readings measured at 570 nm with BSA were subtracted from ECM protein readings. Results from eight experiments performed in triplicate are expressed as the ratio of absorbances measured with miRNA-1b and -2b cells on values obtained with miRNA-Neg cells (means ± S.E.M.).
Growth of miRNA clones and miRNA-Neg cells seeded at low density was then measured every 24 hrs over 96 hrs. As shown in Fig. 3B, cell proliferation was not affected by TFPI-2 down-regulation and no significant difference among miRNA-Neg, miRNA-1b and -2b cell clones was detected (P < 0.05). Moreover, cell viability was similar, as trypan blue staining detected no more than 10% dead cells in all experiments.
Down-regulation of TFPI-2 increases lung cancer cell adhesion to extracellular matrix proteins
To explore further the mechanisms by which TFPI-2 is able to regulate cell invasion, we investigated whether cell attachment to different ECM components was affected when this serine protease inhibitor was down-regulated. The results showed increased adhesion to fibronectin, vitronectin, laminin and collagen IV with both miRNA-1b and -2b clones compared to miRNA-Neg cells (Fig. 3C). Attachment to laminin was particularly marked, with a 2.2-fold and a 2-fold increase for miRNA-1b and -2b when compared to miRNA-Neg cells, respectively, and to collagen IV, with a 1.8-fold and a 1.7-fold for miRNA-1b and -2b, respectively. In contrast, adhesion to collagen I was similar for all cells and no significant differences were observed between miRNA clones and miRNA-Neg cells.
TFPI-2 down-regulation modulates integrin expression on cell surface
To investigate whether the expression of integrins on cell surface could mediate cell-matrix adhesion and thus invasion process, we determined the integrin profiles of cells using various α and β subunit antibodies. Our data clearly demonstrated that down-regulation of TFPI-2 could promote expression of α1 subunit (P < 0.01) while β4 and α5β1 were significantly decreased (P < 0.01) (Fig. 4). In contrast, the expression of the other α and β integrins remained unchanged in miRNA-Neg, mi-RNA-1b and -2b clones.
Fig 4.
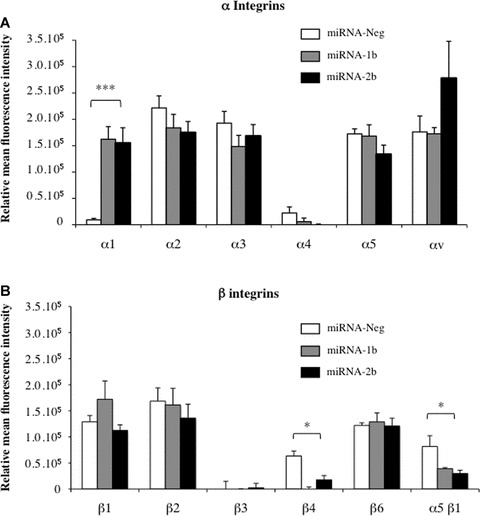
Characterization of cell surface integrins affected by TFPI-2 down-regulation in non-small lung cancer cells. (A) α integrin and (B) β integrin-mediated cell adhesion of tumour cells (3 × 106) transfected with pre-miRNA targeting TFP-2. The fluorescence of CyQuant GR dye evaluating tumour cells adhering to anti-human monoclonal antibody was measured at 535 nm and expressed in relative fluorescence units. Results represent the mean ± S.E.M. of two experiments performed in duplicate. (*P < 0.05, compared with the miRNA-Neg clone, Student t-test.)
Impact of TFPI-2 down-regulation on matrix metalloproteinase expression in lung cancer cells
Levels of MMP-1, -2, -3, -7, -9, -13, -14 and EMMPRIN transcripts were quantified in miRNA-Neg, miRNA-1b and -2b clones using RT and real-time PCR (Fig. 5A). Varying amounts of MMP transcripts were quantified in all cell clones, with fewer than 10 copies for MMP-9, -13 and -14 to more than 105 copies for MMP-1 and EMMPRIN. A 5.5-fold and an 8.6-fold increase in MMP-1 transcripts were found for miRNA-1b and -2b clones, respectively, when compared to miRNA-Neg cells (P < 0.001). Similar results were observed with MMP-3 since mRNA levels were significantly increased (P < 0.01) in miRNA-1b and -2b cell clones (11.3-fold and 10.4-fold, respectively) compared to the miRNA-Neg clone. Immunocytochemical staining also clearly showed that this increase in MMP-1 and -3 transcript levels was associated with enhanced protein expression (Fig. 5B). No significant differences were found in mRNA expression for MMP-9, -13, -14 and EMM-PRIN in either miRNA-1b or -2b compared to the control clone (Fig. 3A). In contrast, MMP-7 and MMP-2 transcript levels were significantly decreased in miRNA-1b (P < 0.01) and -2b (P < 0.001 and P < 0.01, respectively) clones. As shown in Fig. 5(B), MMP-7 protein expression was slightly decreased in both miRNA clones.
Fig 5.
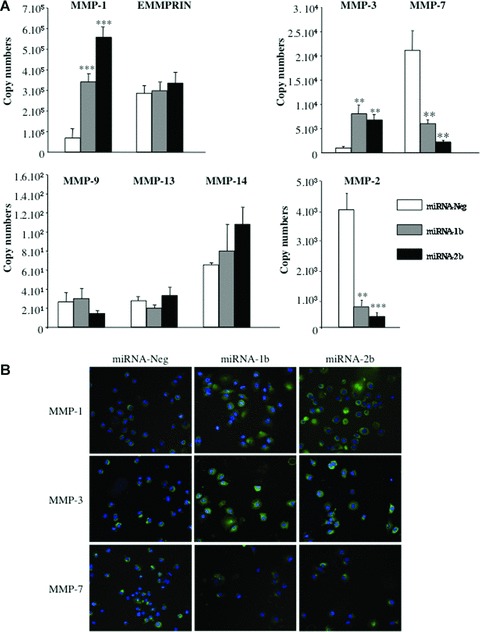
Effects of TFPI-2 down-regulation on metalloproteinase and EMMPRIN in non-small lung cancer cells. (A) MMP and EMMPRIN transcript levels were quantified using real-time RT-PCR in lung cancer cells stably transfected with miRNA-1b and -2b targeting TFPI-2 mRNA and in miRNA-Neg clone cells. The copy numbers of each transcript were normalized to 106 copies of β-actin. Results represent the mean ± S.E.M. from four independent mRNA extractions and real-time RT-PCR performed in triplicate. (*P < 0.05, compared with the miRNA-Neg clone; **P < 0.001; ***P < 0.0001, Student t-test). (B) Immunofluorescence staining of miRNA-1b, -2b and miRNA-Neg clones using monoclonal antibodies against MMP-1, -3 and -7 (original magnification, ×40).
Analysis of signalling pathways
To determine whether various signalling pathways were activated in TFPI-2 down-regulated non-small lung cancer cells, we measured the activities of 10 signal transduction pathways involved in cancer biology using a dual-luciferase reporter assay. As shown in Fig. 6, an increase in relative luciferase activity was observed for extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinases (JNK) and NF-κB pathways in NCI-H460 clones when compared to negative control. In addition, down-regulation of TFPI-2 in miRNA-1b and -2b clones was also associated with a slight increase in NF-κB pathway (P > 0.05) and a significant increase in the ERK and JNK pathways compared to the miRNA-Neg clone (P < 0.05). Other signalling pathway activities remained unchanged in all NCI-H460 clones compared to negative control.
Fig 6.
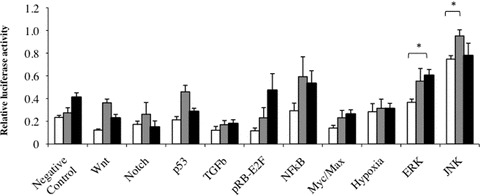
Identification of signal transduction pathway involved when TFPI-2 is down-regulated in lung cancer cells. Using a luciferase pathway reporter array, the firefly luciferase activity of inducible transcription factor-responsive construct transfected in miRNA clones was measured and related to Renilla luciferase activity of constitutively expressing Renilla luciferase construct measured in the same cells for normalizing transfection efficiencies. Results represent the mean ± S.E.M. of experiments performed with miRNA-Neg clone, miRNA-1b and miRNA-2b clone (n= 7). (*P < 0.05, compared with the miRNA-Neg clone, Student t-test).
Effects of conditioned media from TFPI-2 down-regulated non-small lung cancer cells on metalloproteinase expression in lung fibroblasts
In order to examine the effects of soluble factors produced by tumour cells expressing (or not) TFPI-2 on metalloproteinases and EMMPRIN synthesis by fibroblast cells, conditioned serum-free media were obtained from miRNA-Neg, miRNA-1b and -2b clones. Fresh conditioned media equivalent to 107 tumour cells were applied to 106 fibroblasts for 24 hrs. Controls were performed by incubating fibroblasts with non-conditioned medium concentrated 10-fold. MMP-1, -2, -3, -7, -9, -13, -14 and EMMPRIN transcript levels in fibroblast cells, quantified by RT and real-time PCR, were not affected when cells were incubated with concentrated conditioned medium from miRNA-Neg compared to concentrated medium used as control (Fig. 7A). In contrast, a significant increase in MMP-1, -3 and -7 transcript levels were observed when normal lung fibroblasts were stimulated by concentrated conditioned medium obtained from miRNA-1b (P < 0.01) and -2b (P < 0.05) clones. The levels of MMP-2, -9, -13, -14 and EMMPRIN transcripts were also increased with both miRNA 1b and miRNA 2b conditioned media but differences were not significant. As shown in Fig. 7B, the increase in mRNA levels demonstrated by immunofluorescence assay was correlated with the protein expression by fibroblasts, particularly with MMP-1 and MMP-3.
Fig 7.
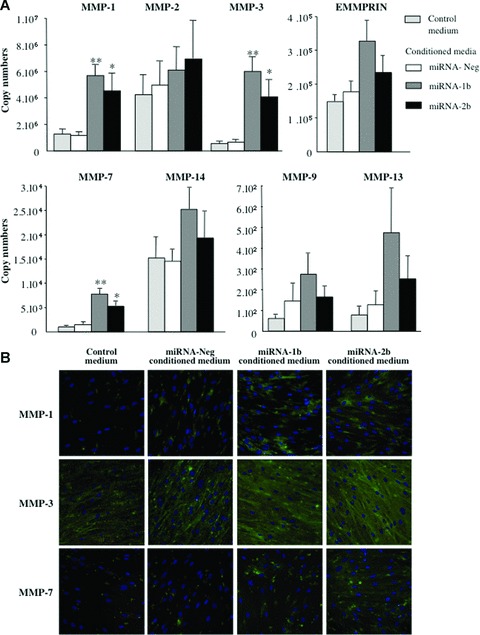
Metalloproteinase and EMMPRIN in fibroblasts cultured with conditioned media from non-small lung cancer cells inactivated for TFPI-2. (A) Normal fibroblasts (106 cells) were grown for 24 hrs in a fresh concentrated conditioned medium from 107 miRNA-1b, -2b, or miRNA-Neg clones. The copy number of each transcript was determined and normalized to 106 copies of β-actin. Data are presented as means ± S.E.M. from four independent mRNA extractions and real-time RT-PCR performed in triplicate. *P < 0.05, cocultured compared with controls; **P < 0.001, Student t-test. (B) Immunofluorescence staining of normal pulmonary fibroblasts using monoclonal antibodies against MMP-1, -3 and -7 (original magnification, ×40).
Discussion
The relationship between TFPI-2 down-regulation, frequently associated with epigenetic changes and tumour aggressiveness, has been well documented in a wide variety of cancers [11, 18]. In agreement with these studies, we have previously demonstrated in non-small lung cancer, representing approximately 85% of all lung cancers, that down-regulation of TFPI-2 and hypermethylation of CpG islands of TFPI-2 promoter are frequently associated with advanced stages of cancer [15]. However, the impact of transcriptional down-regulation of TFPI-2 on tumour cells themselves and on fibroblasts, the major cell component of the tumour microenvironment, is unknown. In the present study, we demonstrated for the first time that the stable TFPI-2 knockdown in non-small lung cancer cells by miRNA increases their adhesion to the ECM laminin and collagen IV and enhances MMP-1 and -3 synthesis, thereby favouring tumour invasion and growth in a nude mice orthotopic model. Furthermore, MMP synthesis was maximized when pulmonary fibroblasts were cocultured with conditioned media from TFPI-2-down-regulated non-small lung cancer cells. These findings raise the possibility that TFPI-2 down-regulation in cancer cells could affect the progression of non-small lung cancer and therefore have important clinical implications.
We showed in this study that TFPI-2 transcripts could be significantly down-regulated in human non-small cell lung cancer cells using artificial microRNA technology. Consistent with this mRNA degradation, both Western blotting and immunofluores-cence assays demonstrated a strong decrease in TFPI-2 protein, mainly when more than 90% of mRNA transcripts were inhibited. The stable targeting and degradation of complementary transcripts by artificial miRNA is a highly specific approach for effective gene silencing [26]. However, miRNA can also reduce the levels of other target transcripts, as demonstrated with miR-21 that regulates several genes including RECK, TIMP-3, PTEN promoting invasiveness of cancer cells [27]. In order to assess the specificity of TFPI-2 knockdown, we transfected non-small lung cancer cells with recombinant plasmids encoding pre-miRNA targeting two different regions of the 3′ UTR of the human TFPI-2 transcript. The specificity of mature miRNA was verified in all functional studies by using a non-silencing miRNA showing no known homology to mammalian genes and called miRNA-Neg. The stability of down-regulation was demonstrated by the measurement of EmGFP fluorescence in clone cells and the levels of TFPI-2 transcripts.
Our results demonstrated that stable TFPI-2 knockdown, as we previously demonstrated with transient RNA interference [23], was associated with a 2-fold increase in invasive ability of NCI-H460 cells through basement membrane components, whereas cell migration through polycarbonate filters was less affected. Our findings are consistent with previous reports demonstrating that the down-regulation of TFPI-2 using an antisense oligonucleotide strategy increased the invasive ability of A549 lung cancer cells in a Matrigel™ invasion assay and the migration of cells from spheroids [28]. Other groups have shown similar results in cancer cell lines derived from melanoma, prostate cancer, glioblastoma and meningioma, and showed that overexpression of TFPI-2 reduced tumour cell invasion [12, 13, 29, 30]. This increased invasion when TFPI-2 was down-regulated cannot be explained by enhanced proliferation as similar cell growth was observed in both miRNA-1b and -2b clone cells and not in miRNA-Neg cells. Moreover, cell viability was similar in all clone cells. We subsequently investigated whether cell adhesion to the ECM might explain the invasive ability of TFPI-2-inactivated lung cancer cells. Our experiments showed that down-regulation of TFPI-2 significantly increased the attachment of miRNA clone cells to ECM proteins, and this was particularly marked for laminin and collagen IV compared to controls. Integrin-mediated cell attachment to collagen IV and laminin mainly depends on the α1 and β1 subunits [31]. Our results indicated that a regulation in α1 integrin subunit on TFPI-2 down-regulation non-small lung cancer cell surface could mediate dynamic linkages between tumour cells and the ECM, affecting a cell's ability to migrate. In addition, the decrease in α5β1 expression, which is a receptor for fibronectin, could also modulate cell adhesion to this ECM component regulating cell-matrix adhesion and migration as suggested in a recent work [32]. Moreover, the invasive behaviour of NCI-H460 cells down-regulated for TFPI-2 was associated with increased MMP-1 and -3 expression. Indeed, MMP-1 is involved in tumour growth, cell invasion and metastasis [33, 34] and elevated expression of MMP-1 has been reported to be associated with poor prognosis in aggressive tumours [35]. Moreover, MMP-1 silencing has been shown to be correlated with decreased ability of chondrosarcoma cells to invade a type I collagen matrix [36] and melanoma cells to form metastases in mice [37]. Secretion of MMP-3 also correlates closely with tumorigenicity and invasiveness of human astrocy-toma cells [38], and migration of muscle cells was reduced when expression of MMP-3 was knocked down using siRNA [39]. The enhanced transcriptional activity of MMP observed in our study may be explained by activation of the mitogen-activated protein kinase (MAPK) pathway involving Ras/Raf/MAPK/extracellular signal-regulated kinase (ERK). Indeed, we demonstrated in this report that inactivation of TFPI-2 in miRNA-1b and -2b NCI-H460 tumour cells activated ERK and JNK signalling and could thus contribute to increase MMP-1 and MMP-3 expression or reduce MMP-2 gene transcription. This result is in agreement with recent report showing in high grade gliomas that loss in LG1, a tumour metastasis suppressor gene, down-regulate MMP-1 and MMP-3 through the PI3K/ERK pathway [40]. The ERK signalling pathway has been also implicated in the activation of many other MMP [41, 42] and it has been shown that down-regulation of this pathway inhibits migration and invasion of tumour cells [43, 44]. As observed with MMP-2, MMP-7 transcripts and protein were lower in NCI-H460 cells expressing miRNA-1b and -2b than in control cells expressing miRNA-Neg. Down-regulation of TFPI-2 could activate signalling pathways reducing MMP-7 transcription. A previous study demonstrated that type 1 insulin-like growth factor receptor could regulate MMP-2 expression by PI3K/Akt and Raf/ERK pathways that transmit opposing signals [45]. In addition, media from TFPI-2-inactivated tumour cells could induce other sig-nalling pathways increasing MMP-7 expression in fibroblasts.
The tumour microenvironment is important for tumour progression, mainly via interactions between tumour cells and fibroblasts. We therefore demonstrated that soluble factors produced by tumour cells enhanced MMP-1, -3, -7 mRNA and protein expression by fibroblasts. Moreover, this increase was more potent when fibroblasts were incubated with media from TFPI-2-inactivated tumour cells. Tumour cells utilize MMP produced by neighbouring fibroblasts rather than by tumour cells themselves for tumour progression, invasion and metastasis [46]. Tumour cells stimulate MMP production via soluble factors such as cytokines and epider-mal growth factor (EGF), fibroblast growth factor (FGF) and tumour necrosis factor-α (TNF-α). Studies have shown that FGF induces transcriptional expression of MMP-1, -2, -3, -9 by fibrob-lasts [47], and that TNF-α stimulates MMP-1 and MMP-3 mRNA expression by fibroblasts [48, 49]. Moreover, EGF can also enhance the transcriptional activity of EMMPRIN [50], a cell surface glycoprotein that stimulates MMP and vascular endothelial growth factor (VEGF) synthesis of both tumour and stromal cells [51]. Moreover, EMMPRIN also exists in soluble form released from either the cell surface via shedding of microvesicles or after proteolytic cleavage by MMP generating EMMPRIN lacking the carboxyl terminus [52]. Protein analyses of conditioned media from tumour cells expressing TFPI-2 (or not) should be compared in order to identify the proteins involved in the increase in MMP expression observed in our coculture study.
In conclusion, the present study provides evidence that inactivation of TFPI-2 synthesis might promote tumour invasion by a mechanism dependent on regulation of MMP-1, -2, -3, -7 and of ERK signalling pathway. Moreover, soluble factors produced by lung cancer cells may contribute to MMP synthesis by fibroblasts from the microenvironment, thus enhancing tumour progression and formation of metastases.
Acknowledgments
We are grateful to Walter Kisiel (Department of Pathology, University of New Mexico, Health Sciences Center, Albuquerque, NM, USA) for kindly providing the anti-TFPI-2 rabbit polyclonal antibody. We thank Alain Le Pape for hepful discussion on this work. We also particularly thank Julien Sobilo, Maryline Lemée, Stéphanie Rétif and Sophie Hamard for excellent technical assistance and Doreen Raine for editing the English text. This study was supported by the ‘Ligue Contre le Cancer’ and the ‘Région Centre’ (THERICAPT project).
References
- 1.Axelrod R, Axelrod DE, Pienta KJ. Evolution of cooperation among tumor cells. Proc Natl Acad Sci USA. 2006;103:13474–9. doi: 10.1073/pnas.0606053103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stamenkovic I. Extracellular matrix remodelling: the role of matrix metalloproteinases. J Pathol. 2003;200:448–64. doi: 10.1002/path.1400. [DOI] [PubMed] [Google Scholar]
- 3.Vihinen P, Kahari VM. Matrix metallopro-teinases in cancer: prognostic markers and therapeutic targets. Int J Cancer. 2002;99:157–66. doi: 10.1002/ijc.10329. [DOI] [PubMed] [Google Scholar]
- 4.Noel A, Jost M, Maquoi E. Matrix metalloproteinases at cancer tumor-host interface. Semin Cell Dev Biol. 2008;19:52–60. doi: 10.1016/j.semcdb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Bjorklund M, Koivunen E. Gelatinase-mediated migration and invasion of cancer cells. Biochim Biophys Acta. 2005;1755:37–69. doi: 10.1016/j.bbcan.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Rao CN, Mohanam S, Puppala, et al. Regulation of ProMMP-1 and ProMMP-3 activation by tissue factor pathway inhibitor-2/matrix-associated serine pro-tease inhibitor. Biochem Biophys Res Commun. 1999;255:94–8. doi: 10.1006/bbrc.1999.0153. [DOI] [PubMed] [Google Scholar]
- 7.Chand HS, Foster DC, Kisiel W. Structure, function and biology of tissue factor pathway inhibitor-2. Thromb Haemost. 2005;94:1122–30. doi: 10.1160/TH05-07-0509. [DOI] [PubMed] [Google Scholar]
- 8.Rao CN, Liu YY, Peavey CL, et al. Novel extracellular matrix-associated serine pro-teinase inhibitors from human skin fibroblasts. Arch Biochem Biophys. 1995;317:311–4. doi: 10.1006/abbi.1995.1168. [DOI] [PubMed] [Google Scholar]
- 9.Iochmann S, Reverdiau-Moalic P, Hube F, et al. Demonstration of inducible TFPI-2 mRNA synthesis in BeWo and JEG-3 tro-phoblast cells using a competitive RT-PCR. Thromb Res. 2002;105:217–23. doi: 10.1016/s0049-3848(02)00018-x. [DOI] [PubMed] [Google Scholar]
- 10.Hube F, Reverdiau P, Iochmann S, et al. Demonstration of a tissue factor pathway inhibitor 2 messenger RNA synthesis by pure villous cytotrophoblast cells isolated from term human placentas. Biol Reprod. 2003a;68:1888–94. doi: 10.1095/biolreprod.102.011858. [DOI] [PubMed] [Google Scholar]
- 11.Sierko E, Wojtukiewicz MZ, Kisiel W. The role of tissue factor pathway inhibitor-2 in cancer biology. Semin Thromb Hemost. 2007;33:653–9. doi: 10.1055/s-2007-991532. [DOI] [PubMed] [Google Scholar]
- 12.Konduri SD, Tasiou A, Chandrasekar N, et al. Role of tissue factor pathway inhibitor-2 (TFPI-2) in amelanotic melanoma (C-32) invasion. Clin Exp Metastasis. 2000;18:303–8. doi: 10.1023/a:1011085820250. [DOI] [PubMed] [Google Scholar]
- 13.Rao CN, Lakka SS, Kin Y, et al. Expression of tissue factor pathway inhibitor 2 inversely correlates during the progression of human gliomas. Clin Cancer Res. 2001;7:570–6. [PubMed] [Google Scholar]
- 14.Wojtukiewicz MZ, Sierko E, Zimnoch L, et al. Immunohistochemical localization of tissue factor pathway inhibitor-2 in human tumor tissue. Thromb Haemost. 2003;90:140–6. [PubMed] [Google Scholar]
- 15.Rollin J, Iochmann S, Blechet C, et al. Expression and methylation status of tissue factor pathway inhibitor-2 gene in non-small-cell lung cancer. Br J Cancer. 2005;92:775–83. doi: 10.1038/sj.bjc.6602298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong CM, Ng YL, Lee JM, et al. Tissue factor pathway inhibitor-2 as a frequently silenced tumor suppressor gene in hepato-cellular carcinoma. Hepatology. 2007;45:1129–38. doi: 10.1002/hep.21578. [DOI] [PubMed] [Google Scholar]
- 17.Hube F, Reverdiau P, Iochmann S, et al. Transcriptional silencing of the TFPI-2 gene by promoter hypermethylation in choriocarcinoma cells. Biol Chem. 2003b;384:1029–34. doi: 10.1515/BC.2003.115. [DOI] [PubMed] [Google Scholar]
- 18.Konduri SD, Srivenugopal KS, Yanamandra N, et al. Promoter methyla-tion and silencing of the tissue factor pathway inhibitor-2 (TFPI-2), a gene encoding an inhibitor of matrix metalloproteinases in human glioma cells. Oncogene. 2003;22:4509–16. doi: 10.1038/sj.onc.1206695. [DOI] [PubMed] [Google Scholar]
- 19.Rao CN, Segawa T, Navari JR, et al. Methylation of TFPI-2 gene is not the sole cause of its silencing. Int J Oncol. 2003;22:843–8. [PubMed] [Google Scholar]
- 20.Nobeyama Y, Okochi-Takada E, Furuta J, et al. Silencing of tissue factor pathway inhibitor-2 gene in malignant melanomas. Int J Cancer. 2007;121:301–7. doi: 10.1002/ijc.22637. [DOI] [PubMed] [Google Scholar]
- 21.Ohtsubo K, Watanabe H, Okada G, et al. A case of pancreatic cancer with formation of a mass mimicking alcoholic or autoim-mune pancreatitis in a young man. Possibility of diagnosis by hypermethyla-tion of pure pancreatic juice. JOP. 2008;9:37–45. [PubMed] [Google Scholar]
- 22.Sprecher CA, Kisiel W, Mathewes S, et al. Molecular cloning, expression, and partial characterization of a second human tissue-factor-pathway inhibitor. Proc Natl Acad Sci USA. 1994;91:3353–7. doi: 10.1073/pnas.91.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iochmann S, Blechet C, Chabot V, et al. Transient RNA silencing of tissue factor pathway inhibitor-2 modulates lung cancer cell invasion. Clin Exp Metastasis. 2009;26:457–67. doi: 10.1007/s10585-009-9245-z. [DOI] [PubMed] [Google Scholar]
- 24.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 25.Gagnadoux F, Pape AL, Lemarie E, et al. Aerosol delivery of chemotherapy in an orthotopic model of lung cancer. Eur Respir J. 2005;26:657–61. doi: 10.1183/09031936.05.00017305. [DOI] [PubMed] [Google Scholar]
- 26.Liang H, Li WH. MicroRNA regulation of human protein protein interaction network. RNA. 2007;13:1402–8. doi: 10.1261/rna.634607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakka SS, Konduri SD, Mohanam S, et al. in vitro modulation of human lung cancer cell line invasiveness by antisense cDNA of tissue factor pathway inhibitor-2. Clin Exp Metastasis. 2000;18:239–44. doi: 10.1023/a:1006755223357. [DOI] [PubMed] [Google Scholar]
- 29.Konduri SD, Tasiou A, Chandrasekar N, et al. Overexpression of tissue factor pathway inhibitor-2 (TFPI-2), decreases the invasiveness of prostate cancer cells in vitro. Int J Oncol. 2001;18:127–31. [PubMed] [Google Scholar]
- 30.Kondraganti S, Gondi CS, Gujrati M, et al. Restoration of tissue factor pathway inhibitor inhibits invasion and tumor growth in vitro and in vivo in a malignant meningioma cell line. Int J Oncol. 2006;29:25–32. [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay NK, Gilchrist D, Gordon GJ, et al. Integrin-dependent protein tyro-sine phosphorylation is a key regulatory event in collagen-IV-mediated adhesion and proliferation of human lung tumor cell line, Calu-1. Ann Thorac Surg. 2004;78:450–7. doi: 10.1016/j.athoracsur.2004.01.042. [DOI] [PubMed] [Google Scholar]
- 32.Chen Q, Meng LH, Zhu CH, et al. ADAM15 suppresses cell motility by driving integrin alpha5beta1 cell surface expression via Erk inactivation. Int J Biochem Cell Biol. 2008;40:2164–73. doi: 10.1016/j.biocel.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 33.Sossey-Alaoui K, Ranalli TA, Li X, et al. WAVE3 promotes cell motility and invasion through the regulation of MMP-1, MMP-3, and MMP-9 expression. Exp Cell Res. 2005;308:135–45. doi: 10.1016/j.yexcr.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Pulukuri SM, Rao JS. Matrix metallopro-teinase-1 promotes prostate tumor growth and metastasis. Int J Oncol. 2008;32:757–65. [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng S, Tada M, Hida Y, et al. High MMP-1 mRNA expression is a risk factor for disease-free and overall survivals in patients with invasive breast carcinoma. J Surg Res. 2008;146:104–9. doi: 10.1016/j.jss.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 36.Jiang X, Dutton CM, Qi WN, et al. siRNA mediated inhibition of MMP-1 reduces invasive potential of a human chondrosar-coma cell line. J Cell Physiol. 2005;202:723–30. doi: 10.1002/jcp.20162. [DOI] [PubMed] [Google Scholar]
- 37.Blackburn JS, Rhodes CH, Coon CI, et al. RNA interference inhibition of matrix met-alloproteinase-1 prevents melanoma metastasis by reducing tumor collagenase activity and angiogenesis. Cancer Res. 2007;67:10849–58. doi: 10.1158/0008-5472.CAN-07-1791. [DOI] [PubMed] [Google Scholar]
- 38.Mercapide J, Lopez De Cicco R, Castresana JS, et al. Stromelysin-1/matrix metalloproteinase-3 (MMP-3) expression accounts for invasive properties of human astrocytoma cell lines. Int J Cancer. 2003;106:676–82. doi: 10.1002/ijc.11286. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura T, Nakamura K, Kishioka Y, et al. Inhibition of matrix metallopro-teinases suppresses the migration of skeletal muscle cells. J Muscle Res Cell Motil. 2008;29:37–44. doi: 10.1007/s10974-008-9140-2. [DOI] [PubMed] [Google Scholar]
- 40.Kunapuli P, Kasyapa CS, Hawthorn L, et al. LGI1, a putative tumor metastasis suppressor gene, controls in vitro invasiveness and expression of matrix metalloproteinases in glioma cells through the ERK1/2 pathway. J Biol Chem. 2004;279:23151–7. doi: 10.1074/jbc.M314192200. [DOI] [PubMed] [Google Scholar]
- 41.Huntington JT, Shields JM, Der CJ, et al. Overexpression of collagenase 1 (MMP-1) is mediated by the ERK pathway in invasive melanoma cells: role of BRAF mutation and fibroblast growth factor signaling. J Biol Chem. 2004;279:33168–76. doi: 10.1074/jbc.M405102200. [DOI] [PubMed] [Google Scholar]
- 42.Shin SY, Choi HY, Ahn BH, et al. Phospholipase Cgamma1 stimulates transcriptional activation of the matrix metalloproteinase-3 gene via the protein kinase C/Raf/ERK cascade. Biochem Biophys Res Commun. 2007;353:611–6. doi: 10.1016/j.bbrc.2006.12.064. [DOI] [PubMed] [Google Scholar]
- 43.Shih YW, Shieh JM, Wu PF, et al. Alpha-tomatine inactivates PI3K/Akt and ERK signaling pathways in human lung adenocarcinoma A549 cells: effect on metastasis. Food Chem Toxicol. 2009;47:1985–95. doi: 10.1016/j.fct.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 44.Yue W, Sun Q, Landreneau R, et al. Fibulin-5 suppresses lung cancer invasion by inhibiting matrix metalloproteinase-7 expression. Cancer Res. 2009;69:6339–46. doi: 10.1158/0008-5472.CAN-09-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang D, Bar-Eli M, Meloche S, et al. Dual regulation of MMP-2 expression by the type 1 insulin-like growth factor receptor: the phosphatidylinositol 3-kinase/Akt and Raf/ERK pathways transmit opposing signals. J Biol Chem. 2004;279:19683–90. doi: 10.1074/jbc.M313145200. [DOI] [PubMed] [Google Scholar]
- 46.Kanekura T, Chen X, Kanzaki T. Basigin (CD147) is expressed on melanoma cells and induces tumor cell invasion by stimulating production of matrix metallopro-teinases by fibroblasts. Int J Cancer. 2002;99:520–8. doi: 10.1002/ijc.10390. [DOI] [PubMed] [Google Scholar]
- 47.Lo J, Hurta RA. Over-expression of K-FGF or bFGF results in altered expression of matrix metalloproteinases: correlations with malignant progression and cellular invasion. Cell Biol Int. 2002;26:319–25. doi: 10.1006/cbir.2001.0858. [DOI] [PubMed] [Google Scholar]
- 48.Domeij H, Yucel-Lindberg T, Modeer T. Signal pathways involved in the production of MMP-1 and MMP-3 in human gingival fibroblasts. Eur J Oral Sci. 2002;110:302–6. doi: 10.1034/j.1600-0722.2002.21247.x. [DOI] [PubMed] [Google Scholar]
- 49.Fang Q, Liu X, Al-Mugotir M, et al. Thrombin and TNF-alpha/IL-1beta synergistically induce fibroblast-mediated collagen gel degradation. Am J Respir Cell Mol Biol. 2006;35:714–21. doi: 10.1165/rcmb.2005-0026OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menashi S, Serova M, Ma L, et al. Regulation of extracellular matrix metalloproteinase inducer and matrix metallopro-teinase expression by amphiregulin in transformed human breast epithelial cells. Cancer Res. 2003;63:7575–80. [PubMed] [Google Scholar]
- 51.Tang Y, Nakada MT, Kesavan P, et al. Extracellular matrix metalloproteinase inducer stimulates tumor angiogenesis by elevating vascular endothelial cell growth factor and matrix metalloproteinases. Cancer Res. 2005;65:3193–9. doi: 10.1158/0008-5472.CAN-04-3605. [DOI] [PubMed] [Google Scholar]
- 52.Nabeshima K, Iwasaki H, Koga K, et al. Emmprin (basigin/CD147): matrix metalloproteinase modulator and multifunctional cell recognition molecule that plays a critical role in cancer progression. Pathol Int. 2006;56:359–67. doi: 10.1111/j.1440-1827.2006.01972.x. [DOI] [PubMed] [Google Scholar]


