Abstract
Chronic wounds – as defined by the World Union of Wound Healing Societies (WUWHS) – are a considerable worldwide health care expense and impair quality of life. In order for chronic wounds to heal, these wounds must be transformed to a more acute state to begin the healing process. Topical negative pressure (TNP) with reticulated open cell foam (ROCF) is known to promote healing in certain types of chronic wounds. However, little is known about changes at the cellular or molecular level in wounds under various treatments, especially under the physical forces induced to tissue by TNP. In the current study, chronic wound samples were obtained during routine wound debridements prior to treatment and 7–12 days after initiating TNP with a continuous setting at –125 mmHg. Whole genome transcriptome microarray analyses were performed on samples to better understand how TNP with ROCF affects these types of wounds. It was found that more genes were expressed following TNP with ROCF as compared to before therapy and to normal, non-wounded tissue. In this study, we show that TNP with ROCF transforms the chronic wound from its inflammation (non-healing) state into more of a progressive, healing phenotype from a molecular point of view with expression of genes that are commonly associated with these terms.
Keywords: topical negative pressure, reticulated open cell foam, vacuum assisted closure therapy, microarray, quantitative real time PCR, transcriptome analysis
Introduction
It has been well established that topical negative pressure manifolded with reticulated open cell foam (TNP/ROCF) using Vacuum Assisted Closure Therapy (V.A.C.® Therapy; Kinetic Concepts, Inc. [KCI] San Antonio, TX, USA) has been used successfully to treat chronic wounds in the clinical setting [1–5]. It is indicated for acute wounds and especially for chronic, non-healing or problematic wounds [6], and has been shown to effectively treat recalcitrant wounds in several randomized controlled trials [7–15]. However, the exact mechanism of action for this therapy has not been fully elucidated.
Acute wounds normally progress through the healing cascade of haemostasis, inflammation, proliferation and remodelling in a timely manner [16]. Chronic wounds first begin as an acute wound; however, they become trapped in a prolonged inflammatory phase of the wound healing cascade [17–19]. These chronically inflamed wounds display classic signs of inflammation, such as redness, swelling, pain and secretion of wound fluid for an extended period of time [20]. At the cellular level, cells in a senescent state may contribute to the chronic wound phenotype [21–23]. It has been shown that fibroblasts in chronic wounds have impaired responses to growth hormone due to the presence of increased numbers of senescent cells [24]. These senescent cells need to be cleared and the wound bed properly prepared before healing of a chronic wound can occur. Therefore, chronic wounds should be re-activated to a more acute wound state in order to resume the healing cascade and prepare the wound bed for eventual closure either by secondary wound healing, surgical closure or reconstructive methods such as flap transfer. V.A.C.® Therapy (TNF/ROCF) has been clinically proven to prepare the wound bed in these types of chronic cases [2, 25–30]. In this study the end-point of this investigation was not to determine if wounds will heal better or not, because with this type of wounds, that is radically debrided and then covered with viable tissue by means of microsurgical transplantation, wound healing mechanisms are not comparable to classical wound healing models or studies that investigate healing phenomena in a tissue that remains and serves as a basis to be reepithelialized. The aim of this study was solely to investigate if TNP does have an effect on a cellular or molecular level or not which has not been investigated so far.
We suggested that TNP/ROCF maintains cells in a metabolically active phenotype following debridement, thereby facilitating wound healing. This prospective clinical study used whole genome microarrays to track changes in gene expression during the treatment of chronic wounds. We compared gene expression in tissue from wounds following TNP/ROCF to tissue prior to TNP/ROCF, as well as to normal skin. The question how the transcriptomes are induced from a TNP standpoint may best be argued by both microdeformation and changes in tissue oxygen gradients. Previous studies from our group on the alteration of Hypoxia-Inducible-Factor (HIF)-1α had shown that oxygen gradients are changed in tissue treated with TNP and certainly this would affect gene expression. Others have shown that many genes are up and down-regulated in fibroblast cultures in response to stretch. These genes are associated with pathways that could be involved in wound healing. Therefore we suggest these two mechanisms (oxygen gradients and microdeformational stretch) associated with TNP therapy may be stimulating changes seen in gene expression [31–33].
Materials and methods
Patients
All patients signed an informed consent and the protocol was approved by the ethics committee of the Friedrich-Alexander-University Erlangen-Nürnberg, Germany (protocol number 3343). Eleven patients (nine male, two female; median age of reporting patients = 71) requiring closure of a severe non-healing, or problematic (exposed vital structures not suitable for wound healing without a surgical flap procedure), or chronic wounds, presented from 2 to 8 weeks were accepted into the study. The wounds were located on the lower extremity with reconstruction planned using free or pedicled flap transfer. Before reconstruction, patients were to receive TNP treatment for wound bed preparation. At two different time-points, each patient had two tissue biopsies collected during routine wound debridements: tissue biopsy from the lower extremity wound site prior to TNP treatment at enrolment date (day 0) and between days 7 and 12 (day 7) after initiating TNP treatment (as seen in Figs 1 and 2 for photographic documentation). Normal non-wound ‘control’ tissue biopsy was also taken from another, non-wound region at ‘day 0’ and ‘day 7’. This means that the control tissue was from the later to be transplanted surgical flap site that was not the treated wound. Therefore we included only appropriate patients who needed a so called delay procedure and who also needed TNP due to the exposure of vital structures with appropriate defects.
Fig 1.
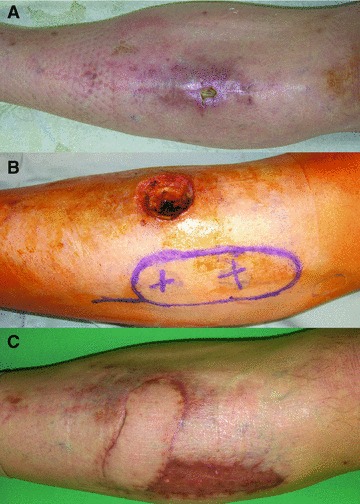
A 50-year-old patient presented with a non-healing chronic wound and osteomyelitis of the tibia in the left pretibial region. The wound was more than 12 months old and 2 × 1 × 1 cm in size (A). The patient was planned to undergo a staged procedure. After debridement, biopsies were taken and TNP was applied. After 5 days of TNP the dressing was removed and the wound bed looked well vascularized (B). Again biopsies of the wound bed and edge were taken and defect coverage was achieved using a distally pedicled Arteria tibialis anterior perforator flap with a size of 9 × 3.5 cm and split skin graft on the donor side. Three months post-operatively stable defect closure was obtained and the patient was mobilized (C).
Fig 2.
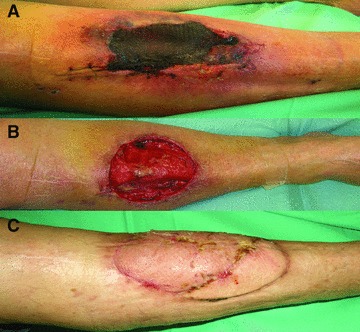
A 70-year-old patient presented with a necrotic soft tissue area of the right ventral lower leg with a size of 5 × 8 cm due to a post-operative compartment syndrome after implantation of a total knee endoprothesis (A). The patient was planned to undergo a staged procedure. After initial debridement, biopsies were taken and TNP was applied. Four weeks after the initial operation, TNP was removed and again biopsies were taken. The wound bed appeared well vascularized (B). Defect coverage was obtained using a free rectus abdominis flap with a skin island. Three months post-operatively stable defect closure was obtained (C).
Treatment of wounds
Prior to patient enrolment in the current study, wounds had been previously treated unsuccessfully with moist saline wound therapy. After the initial sampling of the wound site, wounds were treated with TNP via V.A.C.® Therapy (KCI) for 7 to 12 days. The wound interface material (ROCF) was V.A.C.® GranuFoam® Dressing (KCI), which was covered with V.A.C.® Drape (KCI) and attached to the V.A.C.® Therapy Unit (ATS) via a T.R.A.C.® Pad (KCI). V.A.C.® Therapy was delivered at –125 mmHg on a continuous setting, which is the manufacturer’s recommended default setting.
Tissue samples
On surgery day 0, a 4 mm biopsy of non-wounded skin (‘control day 0’) and debrided wound tissue (‘wound day 0’) was quickly placed into RNAlater® (Ambion, Austin, TX, USA) and stored at –80°C. At surgery days 7–12, a 4 mm biopsy of non-wounded skin (‘control day 7’) and V.A.C.® treated tissue (‘wound day 7’) was quickly removed and stored in RNAlater® at –80°C. Samples were then shipped on dry ice for subsequent preparation of total RNA. Total RNA was isolated from tissue using TRIzol® (Invitrogen, Carlsbad, CA, USA) with clean up modifications to remove DNA using RNeasy columns and a DNase I Kit (Qiagen, Valencia, CA, USA). RNA was stored at –80°C in nuclease-free water (Qiagen). Quality and quantity of RNA was determined using an Experion Automated Electrophoresis system (Bio-Rad, Hercules, CA, USA) and the same RNA samples were divided into aliquots for microarray and TaqMan® quantitative real-time PCR analysis (Applied Biosystems, Foster City, CA, USA). Complete samples of epidermis, dermis and underlying tissue down to the bottom of the deepest wound level were taken. Non-wounded control biopsies were harvested from a point distant to the wound.
Microarray
Due to low RNA quality, high flag numbers or poor scaling, three samples were excluded from microarray analysis; therefore, data from a total of 41 microarrays were compared. Gene expression profiles were generated using the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Santa Clara, CA, USA), which contains over 47,400 transcripts representing approximately 38,500 genes.
Nucleic acid labelling and raw microarray data generation were carried out at Asuragen, Inc. (Austin, TX, USA) according to manufacturer’s protocols. Prior to amplification and labelling, the quality and quantity of total RNA isolated from tissues was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Raw data files were analysed through the Vanderbilt Microarray Shared Resource (Nashville, TN, USA) with the use of Partek® Genomics Suite (St. Louis, MO, USA). Removal of batch effect was employed due to the running of the microarrays on two different dates. Probe signal intensities across microarrays were normalized using robust multichip average. Features with signal/noise values ≥3 and quality flag values <5000 were considered detected and were compared with a P-value of <0.001. Fold change values were calculated from probe intensities.
Validation using TaqMan®-based quantitative real-time PCR gene expression assays
Validation of the microarray data was performed by quantitative real-time PCR. Approximately 1 μg of total RNA of each sample per 100 μl reaction was used to generate cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The TaqMan® assays for the FAM™ labelled gene of interest was duplexed with the VIC® labelled 18S RNA endogenous control assay using a 7500 Fast real-time PCR system (Applied Biosystems). For each sample, three technical replicates per gene were run in a 96-well format plate and on each plate a no-template control was also run in triplicate. Relative quantification analysis was performed with SDS software v. 1.3.1 (Applied Biosystems).
Data analysis
The microarray data analysis was generated using Partek® software (Partek® Inc., St. Louis, MO, USA). To look for genes that were different between wound and control tissue at 1 week, each patient’s wound ‘day 7’ sample was normalized against their own control ‘day 7’ sample (day 7 analysis). To look for genes that were different between wound and control tissue at day 0, each patient’s wound ‘day 0’ sample was normalized against their own control ‘day 0’ sample (day 0 analysis). To look for genes changing only at the wound site, wound day 7 samples were compared to wound day 0 samples (wound analysis). Partek® data were further analysed through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, Redwood City, CA, USA). Multivariate correlation for qPCR and microarray analysis was analysed through JMP Version 7.0 (SAS, Cary, NC, USA). Fold change binning was generated by the use of an Access database (Microsoft, Redmond, WA, USA).
Results
Gene pathways
When looking at raw probe data, P < 0.001, there were a total of 606 probes which were differentially expressed at day 0 (before TNP treatment). Following TNP therapy, there were a total of 851 probes which were differentially expressed. Only 65 probes were common between day 0 and day 7 time-points (Fig. 3). Using Ingenuity for pathway analysis listing and in-depth gene data, it was determined that the networks with the most differentially expressed genes for day 0 (before treatment) were cellular compromise and infection mechanisms. Genes within these networks were most significantly associated (P < 0.05) with cell death, death receptor signalling, oxidative stress response, physiological system development and function. After treatment on day 7, networks with the most significantly differentially expressed genes (P < 0.05) in TNP treated tissue included cellular assembly and organization, cellular function and maintenance, and RNA post-transcriptional modification, cellular growth and proliferation, hair and skin development and tissue development. Genes significantly expressed (P < 0.05) within these networks were most associated with integrin signalling and transforming growth factor β (TGF-β) signalling. Table 1 shows the associated Ingenuity networks and pathways.
Fig 3.
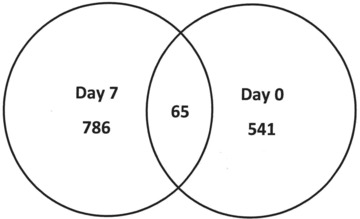
Venn diagram of raw probe data from Partek®. There were a total of 606 probes [541 plus 65] differentially expressed on day 0 and 851 probes [786 plus 65] differentially expressed for day 7 following TNP with ROCF. Of these, 65 probes were commonly expressed between time-points (P < 0.001).
Table 1.
Significant genes associated Ingenuity networks and pathways as categorized for day 0 and day 7 (P < 0.05)
| Top networks | Day 0 | Day 7 |
|---|---|---|
| Cellular compromise | x | |
| Infection mechanism | x | |
| Cellular assembly and organization | x | |
| Cellular function and maintenance | x | |
| RNA post-transcriptional modification | x | |
| Molecular and cellular functions | ||
| Cell death | x | |
| Cellular growth and proliferation | x | |
| Top canonical pathways | ||
| Death receptor signalling | x | |
| Integrin signalling | x | |
| Top toxicological list | ||
| Pro-apoptosis | x | |
| Oxidative stress response | x | |
| TGF-β signalling | x | |
| Physiological system development and function | ||
| Cell-mediated immune response | x | |
| Hair and skin development and function | x | |
| Tissue development | x | |
Gene data were entered into Ingenuity Pathways Analysis biomarker, where genes unique to each data set, regardless of P-value, are compared to each other. As shown in Figure 4 there were a total of 213 unique genes expressed on day 0 (before treatment), 2551 unique genes for day 7 (after TNP treatment) and 211 genes which were common between day 0 and day 7. When gene data were filtered for epidermis genes, there were 102 unique genes for day 0 (before treatment), 1277 for day 7 (after TNP treatment) and there were 113 common genes between day 0 and day 7. This shows that approximately 50% of the unique biomarkers (day 0, day 7 and common) were epidermis related genes. Figure 5 depicts these relationships.
Fig 4.
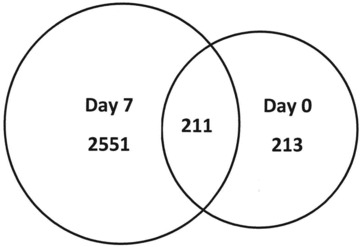
Venn diagram of unique gene data sets from Ingenuity Biomarker analysis tool showing; 2551 unique genes were expressed for day 7 (after TNP/ROCF), whereas only 213 unique genes were expressed for day 0 (before treatment). A total of 211 genes were commonly expressed between days 0 and 7.
Fig 5.
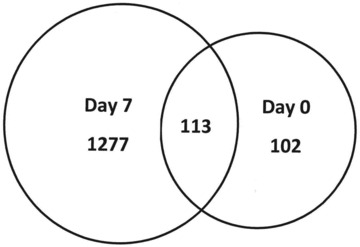
Venn diagram of unique gene data sets from Ingenuity Biomarker analysis tool – biofiltered for human epidermis gene expression only showing; 1277 unique genes were expressed on day 7 (after TNP/ROCF treatment), whereas 102 unique genes were expressed on day 0 (before treatment). A total of 113 epidermis related genes were expressed in common between the two time-points.
Ingenuity Pathways Analysis gene data were next binned for fold change, both up- and down-regulated, from 2 to 10 and greater than 10-fold differences (Table 2). There were more genes differentially expressed from control tissue on day 7 (545 down-regulated; 322 up-regulated) than on day 0 (19 down-regulated; 31 up-regulated) with 88.0% of these binned genes having a fold change of 2–10. Of interest were the top 10 up- and down-regulated genes on day 7 (Table 3). The majority of the down-regulated genes were associated with cell adhesion, whereas the up-regulated genes were associated with macrophage-mediated immunity and the extracellular matrix. When filtered for epidermis genes (data not shown), the majority of the genes were also differentially expressed on day 7 (309 down-regulated; 167 up-regulated) compared to day 0 (11 down-regulated; 15 up-regulated) with 86.8% of these binned genes having a fold change of 2–10.
Table 2.
Fold change bins, up- and down-regulated, for Ingenuity Pathway Analysis – biomarker data on day 0 and day 7. There were more genes exhibiting high-fold change on day 7 than on day 0
| Ingenuity Pathway Analysis – biomarker | |||
|---|---|---|---|
| Day 0 | Day 7 | ||
| ↓ FC | # of genes | ↓ FC | # of genes |
| 2–10 | 19 | 2–10 | 443 |
| >10 | 0 | >10 | 102 |
| Total | 19 | Total | 545 |
| ↑ FC | # of genes | ↑ FC | # of genes |
| 2–10 | 31 | 2–10 | 320 |
| >10 | 0 | >10 | 2 |
| Total | 31 | Total | 322 |
Table 3.
Fold change bins, up and down-regulated, for Ingenuity Pathway Analysis – biomarker data on day 0 and day 7. There were more genes exhibiting high-fold change on day 7 than on day 0
| Top 10 down-regulated genes – day 7 | |||||
|---|---|---|---|---|---|
| Affymetrix ID | Fold change | Entrez gene name | Symbol | Function | Location |
| 209351_at | −265 | Keratin 14 | KRT14 | Cell structure | Cytoplasm |
| 207324_s_at | −190 | Desmocollin 1 | DSC1 | Cell adhesion | Plasma membrane |
| 206400_at | −160 | Lectin, galactoside-binding, soluble, 7B | LGALS7B | Cell adhesion | Unknown |
| 206642_at | −143 | Desmoglein 1 | DSG1 | Cell adhesion | Plasma membrane |
| 205694_at | −112 | Tyrosinase-related protein 1 | TYRP1 | Carbon metabolism; oxygenase; enzyme | Cytoplasm |
| 226926_at | −108 | Dermokine | DMKN | Keratinocyte-secreted peptide | Unknown |
| 206032_at | −102 | Desmocollin 3 | DSC3 | Cell adhesion | Plasma membrane |
| 204855_at | −101 | Serpin peptidase inhibitor, clade B (ovalbumin), member 5 | SERPINB5 | Protease inhibitor | Extracellular space |
| 224650_at | −92 | Mal, T-cell differentiation protein 2 | MAL2 | Membrane traffic transporter | Plasma membrane |
| 201131_s_at | −91 | Cadherin (CDH) 1, type 1, E-cadherin (epithelial) | CDH1 | Cell adhesion | Plasma membrane |
Validation of microarray results by qPCR
Microarray results were validated by quantitative real-time PCR analysis of six individual genes. These genes were chosen for their respective roles in wound healing. For this study, we chose epidermis growth factor receptor, defender against cell death, caspase 9, striatin, fibroblast growth factor 12 and chemokine receptor like 1. There was an 88% correlation between microarray and quantitative real-time PCR results. This percentage may be arbitrarily low because certain selected TaqMan® inventoried gene assays might not have covered all possible gene transcripts for the gene of interest and normalization of data to control tissue. However, the positive correlation between qPCR and microarray data validates the use of microarray data for genomic interpretation.
Discussion
At study day 0, patients presented with problematic non-healing wounds which were clinically not ready for closure and required wound bed optimization with TNP before a necessary plastic surgical coverage was performed to close non-healing structures. Common methods of reconstruction are still necessary when a wound fails to heal or when vital structures are exposed that need tissue coverage. Methods of tissue engineering have been utilized in this context, but despite recent advances in providing in vivo and in vitro assembled vascularized tissue substitutes these techniques are not a clinically available method yet [34–43]. Therefore, a clinical judgment was made to utilize TNP/ROCF to prepare the wound bed for reconstruction using free or pedicled flap transfer. Between 7 and 12 days following TNP/ROCF, a clinical judgment was made that the wounds had progressed to allow for successful defect coverage using flap procedures. This was photographically documented as seen in Figures 1 and 2. Microarray data provide insight into wound environment physiology to allow for better genomic understanding of how a chronic wound progresses to one which could be closed surgically. In this preliminary study, there were more genes actively up- or down-regulated in day 7 tissue after TNP than on day 0 or in normal tissue samples. At day 0, the most common genes represented were for cell death, whereas on day 7, there were more structural genes expressed. At day 0, 213 unique genes were differentially expressed from control whereas there were 2551 genes differentially expressed on day 7. This would suggest that prior to treatment with TNF, gene expression was low and this corresponded to a chronic wound phenotype. Seven days of TNP stimulated unique gene expression 10-fold over day 0, corresponding to a healing wound phenotype. This is reiterated in the top regulated genes for day 7, where down-regulation of cell adhesion genes was occurring whereas extracellular matrix and macrophage-mediated immunity genes were up-regulated.
Based upon Partek® and Ingenuity analysis, following TNP treatment, differentially expressed genes were associated with anabolic processes such as cellular assembly, function and organization. This suggests that TNP was affecting restructuring of the wound at the cellular level to promote healing. This was validated clinically; as the chronic wounds had progressed in a short amount of time to a point where closure could be accomplished using plastic surgery methods, e.g. flap transfer [44, 45]. The increased level of gene expression following TNP/ROCF could be due in part to macro and microstrain. It has been shown previously that TNP imparts 5–20% strain to tissue [46]. This level of strain is consistent with that known to promote cellular proliferation after V.A.C.® Therapy application [46]. Treatment of fibroblasts in a three-dimensional matrix with TNP leads to an increase in cell migration, proliferation and protein synthesis [47, 48], processes that are all important to the formation of granulation tissue. Novel gene expression presumably is required for healing to occur.
In the current study, following treatment with TNP/ROCF clear patterns of gene expression for genes with high-fold change were observed. For example, up-regulated genes with the highest increase in regulation included a variety of extracellular matrix genes. Epiphycan, an extracellular matrix proteoglycan, is up-regulated 51-fold whereas fibronectin is up-regulated 7-fold. Also, genes involved in macrophage signalling were up-regulated. These included CD163 and macrophage scavenger receptor 1. Before wounds can enter the proliferative phase, macrophages must enter the wound and synthesize and release growth factors (M2 phenotype) [49]. Increased expression of macrophage and extracellular membrane genes at day 7 provides evidence of up-regulation of pro-healing anabolism-related genes. A potential limitation of this model may be seen in the fact that the patients serve as their own control. However, because biopsies from the wound margin and the wound under the TNP dressing were compared with tissue biopsies that were not treated at all gives a realistic comparison as to the study aim, which was to determine any potential effect of TNP on the genomic level. Because no other variables other than the exertion of TNP to the tissue were used, this might also be seen as an advantage compared to other wound models with different patients and variable underlying diseases. As for experiments designed to show the acceleration of healing due to TNP we propose this may be done in an animal experiment. In such an experiment one could look at gene expression or some other parameter over time in a delayed healing model and look at time to granulation fill. This type of animal experiment that has yet to be designed would offer additional possibilities to study the genome alteration under various types of TNP and allow for more tissue samples than possible in the human being.
In conclusion, TNP as delivered by V.A.C. ® Therapy helped to prepare chronic wounds for closure. Microarray analysis proved that, on a molecular basis, genes are being regulated much differently between different treatments in the study group. A chronic, non-healing, poorly expressing wound was treated with TNP/ROCF and after treatment of at least 7 days the wound was transformed to a ‘healing phenotype’. Future studies will determine if gene expression differs between chronic wounds treated with different modalities.
Acknowledgments
Preliminary data of this study have been presented at the Drei-Länder-Kongress on VAC Therapy in Zürich, 16 April 2010 and have been printed in the supplementary abstract issue of the Zeitschrift für Wundheilung. This study was supported by a research grant of Kinetic Concepts Inc. to the Friedrich-Alexander-University of Erlangen-Nuernberg (Grant Nr. VACP 2005-17; Erlangen University Nr. Fe 34499008). The study was approved by the Ethical committee Friedrich-Alexander-University of Erlangen-Nuernberg clearance: Nr. 3343. We thank PD Dr. med. Ulrich Kneser for operating the clinical cases presented in Figures 1 and 2.
Disclosures
Kathleen L. Derrick, M.S., and Amy McNulty, Ph.D. are employed at Kinetic Concepts Inc. (KCI) research department. The first and senior authors have served as lecturers for KCI.
Conflict of interest
The authors confirm that there are no conflicts of interest.
References
- 1.Argenta LC, Morykwas MJ. Vacuum-assisted closure: a new method for wound control and treatment: clinical experience. Ann Plast Surg. 1997;38:563–76. [PubMed] [Google Scholar]
- 2.Joseph E, Hamori CA, Bergman S, et al. A prospective, randomized trial of vacuum-assisted closure versus standard therapy of chronic nonhealing wounds. Wounds. 2000;12:60–7. [Google Scholar]
- 3.Short B, Claxton M, Armstrong DG. How to use VAC therapy on chronic wounds. Podiatry Today. 2002;15:48–54. [Google Scholar]
- 4.Sibbald RG, Mahoney J. A consensus report on the use of vacuum-assisted closure in chronic, difficult-to-heal-wounds. Ostomy Wound Manage. 2003;49:52–66. [PubMed] [Google Scholar]
- 5.Venturi ML, Attinger CE, Mesbahi AN, et al. Mechanisms and clinical applications of the vacuum-assisted closure (VAC) device: a review. Am J Clin Dermatol. 2005;6:185–94. doi: 10.2165/00128071-200506030-00005. [DOI] [PubMed] [Google Scholar]
- 6.Kinetic Concepts Incorporated. V.A.C. therapy clinical guidelines. San Antonio: KCI Licensing; 2006. [Google Scholar]
- 7.Armstrong DG, Lavery LA. Negative pressure wound therapy after partial diabetic foot amputation: a multicentre, randomised controlled trial. Lancet. 2005;366:1704–10. doi: 10.1016/S0140-6736(05)67695-7. [DOI] [PubMed] [Google Scholar]
- 8.Braakenburg A, Obdeijn MC, Feitz R, et al. The clinical efficacy and cost effectiveness of the vacuum-assisted closure technique in the management of acute and chronic wounds: a randomized controlled trial. Plast Reconstr Surg. 2006;118:390–7. doi: 10.1097/01.prs.0000227675.63744.af. [DOI] [PubMed] [Google Scholar]
- 9.Eginton MT, Brown KR, Seabrook GR, et al. A prospective randomized evaluation of negative-pressure wound dressings for diabetic foot wounds. Ann Vasc Surg. 2003;17:645–9. doi: 10.1007/s10016-003-0065-3. [DOI] [PubMed] [Google Scholar]
- 10.Etoz A, Ozgenel Y, Ozcan M. The use of negative pressure wound therapy on diabetic foot ulcers: a preliminary controlled trial. Wounds. 2004;16:264–9. [Google Scholar]
- 11.Ford CN, Reinhard ER, Yeh D, et al. Interim analysis of a prospective, randomized trial of vacuum-assisted closure versus the healthpoint system in the management of pressure ulcers. Ann Plast Surg. 2002;49:55–61. doi: 10.1097/00000637-200207000-00009. [DOI] [PubMed] [Google Scholar]
- 12.McCallon SK, Knight CA, Valiulus JP, et al. Vacuum-assisted closure versus saline-moistened gauze in the healing of postoperative diabetic foot wounds. Ostomy Wound Manage. 2000;46:28–34. [PubMed] [Google Scholar]
- 13.Moues CM, Vos MC, van den Bemd GJ, et al. Bacterial load in relation to vacuum-assisted closure wound therapy: a prospective randomized trial. Wound Repair Regen. 2004;12:11–7. doi: 10.1111/j.1067-1927.2004.12105.x. [DOI] [PubMed] [Google Scholar]
- 14.Smith APS, Kieswetter K, Goodwin AL. Negative pressure wound therapy. In: Krasner DL, Rodeheaver G, Sibbald RG, et al., editors. Chronic wound care: a clinical source book for healthcare professionals. Pennsylvania: HMP Communications; 2007. pp. 271–86. [Google Scholar]
- 15.Vuerstaek JD, Vainas T, Wuite J, et al. State-of-the-art treatment of chronic leg ulcers: a randomized controlled trial comparing vacuum-assisted closure (V.A.C.) with modern wound dressings. J Vasc Surg. 2006;44:1029–37. doi: 10.1016/j.jvs.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Diegelmann RF, Evans MC. Wound healing: an overview of acute, fibrotic and delayed healing. Front Biosci. 2004;9:283–9. doi: 10.2741/1184. [DOI] [PubMed] [Google Scholar]
- 17.Schultz GS, Ladwig G, Wysocki A. Extracellular matrix: review of its roles in acute and chronic wounds. World Wide Wounds. 2005 . Available at: http://www.worldwidewounds.com/2005/august/Schultz/Extrace-Matric-Acute-Chronic-Wounds.html. . Accessed January 28, 2009. [Google Scholar]
- 18.Roy S, Biswas S, Khanna S, et al. Characterization of a preclinical model of chronic ischemic wound. Physiol Genomics. 2009;37:211–24. doi: 10.1152/physiolgenomics.90362.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy S, Patel D, Khanna S, et al. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci USA. 2007;104:14472–7. doi: 10.1073/pnas.0706793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnstone CC, Farley A, Hendry C. The physiological basics of wound healing. Nurs Stand. 2005;19:59–66. doi: 10.7748/ns2005.07.19.43.59.c3906. [DOI] [PubMed] [Google Scholar]
- 21.Shelton DN, Chang E, Whittier PS, et al. Microarray analysis of replicative senescence. Curr Biol. 1999;9:939–45. doi: 10.1016/s0960-9822(99)80420-5. [DOI] [PubMed] [Google Scholar]
- 22.Brem H, Stojadinovic O, Diegelmann RF, et al. Molecular markers in patients with chronic wounds to guide surgical debridement. Mol Med. 2007;13:30–9. doi: 10.2119/2006-00054.Brem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charles CA, Tomic-Canic M, Vincek V, et al. A gene signature of nonhealing venous ulcers: potential diagnostic markers. J Am Acad Dermatol. 2008;59:758–71. doi: 10.1016/j.jaad.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding KG, Morris HL, Patel GK. Science, medicine and the future: healing chronic wounds. BMJ. 2002;324:160–3. doi: 10.1136/bmj.324.7330.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans D, Land L. Topical negative pressure for treating chronic wounds. Cochrane Database Syst Rev. 2001;1 doi: 10.1002/14651858.CD001898. : CD001898. [DOI] [PubMed] [Google Scholar]
- 26.Apelgvist J, Armstrong DG, Augustin M, et al. Vacuum assisted closure: recommendations for use. A consensus document. Int Wound J. 2008;5:1–19. doi: 10.1111/j.1742-481X.2008.00537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bovill E, Banwell PE, Teot L, et al. Topical negative pressure wound therapy: a review of its role and guidelines for its use in the management of acute wounds. Int Wound J. 2008;5:511–29. doi: 10.1111/j.1742-481X.2008.00437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horch RE. The development of plastic surgery: retrospective view of 80 years of “Der Chirurg” (The Surgeon) Chirurg. 2009;80:1132–9. doi: 10.1007/s00104-009-1778-9. [DOI] [PubMed] [Google Scholar]
- 29.Horch RE, Dragu A, Lang W, et al. Coverage of exposed bones and joints in critically ill patients: lower extremity salvage with topical negative pressure therapy. J Cutan Med Surg. 2008;12:223–9. doi: 10.2310/7750.2008.07073. [DOI] [PubMed] [Google Scholar]
- 30.Horch RE, Nord D, Augustin M, et al. Economic aspects of surgical wound therapies. Chirurg. 2008;79:518–25. doi: 10.1007/s00104-008-1500-3. [DOI] [PubMed] [Google Scholar]
- 31.Dragu A, Schnurer S, Surmann-Schmitt C, et al. Gene expression analysis of ischemia and reperfusion in human microsurgical free muscle tissue transfer. J Cell Mol Med. 2010 doi: 10.1111/j.1582-4934.2010.01061.x. 10.1111/j.1582-4934.2010.01061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimm A, Dimmler A, Stange S, et al. Expression of HIF-1 alpha in irradiated tissue is altered by topical negative-pressure therapy. Strahlenther Onkol. 2007;183:144–9. doi: 10.1007/s00066-007-1560-1. [DOI] [PubMed] [Google Scholar]
- 33.Mackley JR, Ando J, Herzyk P, et al. Phenotypic responses to mechanical stress in fibroblasts from tendon, cornea and skin. Biochem J. 2006;396:307–16. doi: 10.1042/BJ20060057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arkudas A, Tjiawi J, Saumweber A, et al. Evaluation of blood vessel ingrowth in fibrin gel subject to type and concentration of growth factors. J Cell Mol Med. 2009;13:2864–74. doi: 10.1111/j.1582-4934.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bleiziffer O, Horch RE, Hammon M, et al. T17b murine embryonal endothelial progenitor cells can be induced towards both proliferation and differentiation in a fibrin matrix. J Cell Mol Med. 2009;13:926–35. doi: 10.1111/j.1582-4934.2008.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiegel HC, Kaufmann PM, Bruns H, et al. Hepatic tissue engineering: from transplantation to customized cell-based liver directed therapies from the laboratory. J Cell Mol Med. 2008;12:56–66. doi: 10.1111/j.1582-4934.2007.00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiegel HC, Pryymachuk G, Rath S, et al. Fetal hepatocyte transplantation in a vascularized AV-loop transplantation model in the rat. J Cell Mol Med. 2010;14:267–74. doi: 10.1111/j.1582-4934.2008.00369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horch RE. Future perspectives in tissue engineering. J Cell Mol Med. 2006;10:4–6. doi: 10.1111/j.1582-4934.2006.tb00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horch RE, Kopp J, Kneser U, et al. Tissue engineering of cultured skin substitutes. J Cell Mol Med. 2005;9:592–608. doi: 10.1111/j.1582-4934.2005.tb00491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horch RE, Popescu LM, Vacanti C, et al. Ethical issues in cellular and molecular medicine and tissue engineering. J Cell Mol Med. 2008;12:1785–93. doi: 10.1111/j.1582-4934.2008.00460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutmacher DW, Horch RE, Loessner D, et al. Translating tissue engineering technology platforms into cancer research. J Cell Mol Med. 2009;13:1417–27. doi: 10.1111/j.1582-4934.2009.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polykandriotis E, Euler S, Arkudas A, et al. Regression and persistence: remodelling in a tissue engineered axial vascular assembly. J Cell Mol Med. 2009;13:4166–75. doi: 10.1111/j.1582-4934.2009.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallmichrath J, Stark GB, Kneser U, et al. Epidermal growth factor (EGF) transfection of human bone marrow stromal cells in bone tissue engineering. J Cell Mol Med. 2009;13:2593–601. doi: 10.1111/j.1582-4934.2008.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leffler M, Derrick KL, McNulty A, et al. Übersicht über die molekularbiologischen Grundlagen der V.A.C. Therapie. Zeitschrift f Wundheilung. 2010;15:6–10. [Google Scholar]
- 45.Leffler M, McNulty A, Kneser U, et al. Analyse von Genexpressionsmustern und Intrazellulären Signalwegen in akuten Wunden unter Vakuumbehandlung (V.A.C.®)–Erste Ergebnisse einer Prospektiv-Kontrollierten Klinischen Studie. Zeitschrift f Wundheilung. 2009;14:9–10. [Google Scholar]
- 46.Saxena V, Hwang CW, Huang S, et al. Vacuum-assisted closure: microdeformations of wounds and cell proliferation. Plast Reconstr Surg. 2004;114:1086–96. doi: 10.1097/01.prs.0000135330.51408.97. [DOI] [PubMed] [Google Scholar]
- 47.McNulty AK, Schmidt M, Feeley T, et al. Effects of negative pressure wound therapy on fibroblast viability, chemotactic signaling, and proliferation in a provisional wound (fibrin) matrix. Wound Repair Regen. 2007;15:838–46. doi: 10.1111/j.1524-475X.2007.00287.x. [DOI] [PubMed] [Google Scholar]
- 48.McNulty AK, Schmidt M, Feeley T, et al. Effects of negative pressure wound therapy on cellular energetics in fibroblasts grown in a provisional wound (fibrin) matrix. Wound Repair Regen. 2009;17:192–9. doi: 10.1111/j.1524-475X.2009.00460.x. [DOI] [PubMed] [Google Scholar]
- 49.Brown BN, Valentin JE, Stewart-Akers AM, et al. Macrophage phenotype and remodeling outcomes in response to biologic scaffolds with and without a cellular component. Biomaterials. 2009;30:1482–91. doi: 10.1016/j.biomaterials.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]


