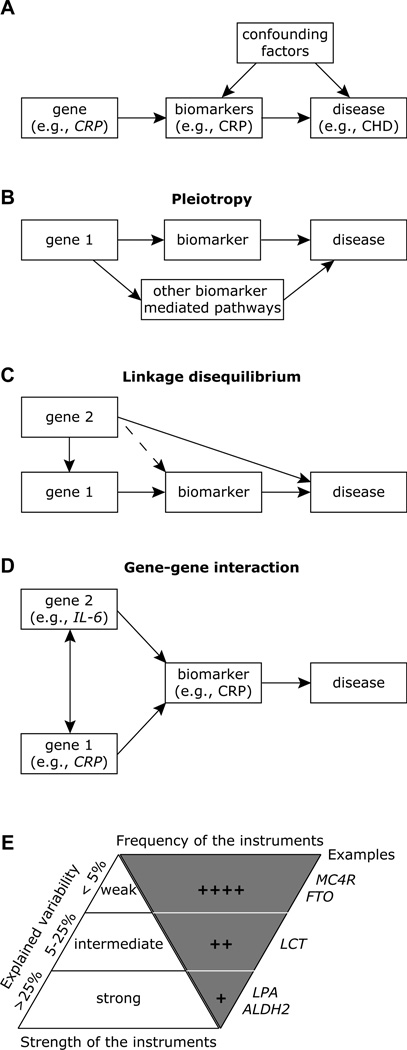
Fig. 3.
Limitations and considerations while applying Mendelian randomization. (A) The CRP genotype as an instrumental variable in Mendelian randomization. The arrows can be considered to represent causal relationships; it is important to note that there is no arrow between CRP gene and the confounders and there is no single arrow between CRP gene and CHD, i.e., there are no other routes in the pictorial between CRP gene and CHD. (B) The problem of pleiotropy. A single gene can influence disease risk through multiple pathways other than the biomaker. This violates the second assumption of Mendelian randomization. (C) Problem of linkage disequilibrium. In this instance, the genetic variant is in linkage disequilibrium with the variant associated with, or causal for, the disease. SNP in Gene 1 is in linkage disequilibrium with another SNP in Gene 2. Here, Gene 2 directly affects the disease level or risk, and hence Gene 1 is not an adequate genetic instrument for MR. The SNP in Gene 1 could be in linkage disequilibrium with a SNP in Gene 2 that is also causal for the biomarker (dashed line). In this instance, if Gene 2 SNP only affects disease via its effects on the same biomarker, there is no violation of the assumptions, and SNP in Gene 1 can still be considered as an instrument in a Mendelian randomization analysis. (D) Problem of gene-gene interaction. Example shown is for the interaction between genetic variants in IL-6 and CRP wherein a polymorphism in IL-6 gene correlates more closely with CRP levels than IL-6 levels. (E) Low explained variability. The Mendelian randomization approach is dependent on the proportion of the variance (explained variability depicted along the left of the upright triangle) of the biomarker explained by the genetic instrument (SNP) for the genes depicted on along the right of the inverted triangle. Abbreviations for genes: MC4R, melanocortin 4 receptor gene; FTO, fast mass and obesity associated gene; CRP, C-reactive protein gene; LCT, lactase gene; LPA, lipoprotein A gene; and ALDH2, aldehyde dehydrogenase gene. Data is adapted from [11].