3.
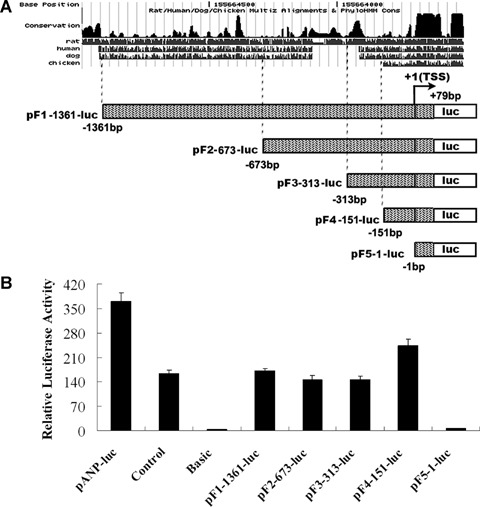
Truncation analysis of promoter regions in the 5′ flanking region of mouse CARK gene. (A) A series of constructs containing various lengths of the 5′ region of the CARK promoter regions based on the conservation between mouse and other mammals using ClustalW program were fused to the luciferase reporter. (B) These constructs were introduced into primary rat cardiomyocytes, HEK293S cells, Hela cells, rat aorta smooth muscle cells and rat cardiac fibroblasts for transient expression assays. After 48 hrs, cells were lysed and luciferase activities were assayed. Firefly luciferase activities expressed had been normalized on the basis of Renilla luciferase activity encoded by the co-transfected control plasmid, pRL-CMV. Data shown are the means ± S. D., n= 3. The results showed that the luciferase activity was detectable only in rat cardiomyocytes. pGL3 basic vector containing no promoter element was used as negative control and pGL3 promoter vector fused with SV40 promoter element was used as positive control. A 2.7-kb long promoter region of ANP gene was sub-cloned into pGL3 basic vector (named as pANP-luc) as positive control.
