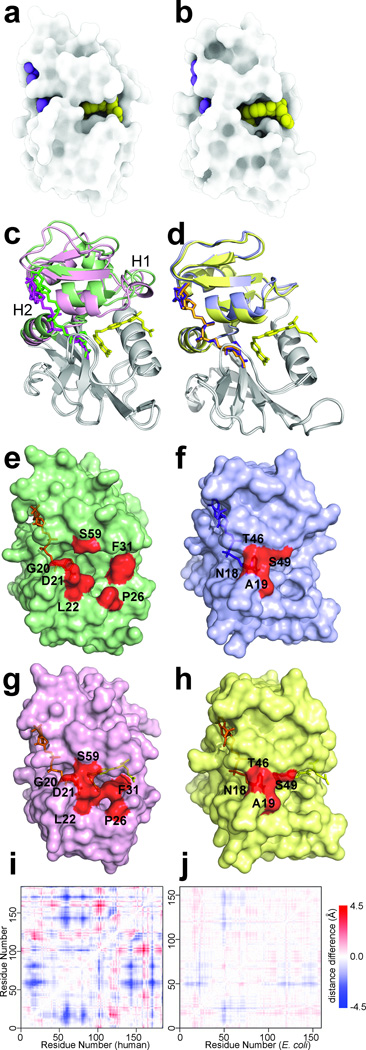
Figure 2. Active site packing and hinge motions in hDHFR.
a,b Surface rendition of hDHFR–NADP+–FOL (a) and ecDHFR:NADP+–FOL (b) generated using only ambient occlusion, a 3D light attenuation calculation where deep pockets render dark and exposed surfaces render light35. c,d Superposition of crystal structures, aligned on the loop subdomain (gray), of hE–NADPH and hE–NADP+–FOL (c) and ecE–NADPH (PDB code: 1RX118) and ecE–NADP+–FOL (PDB code: 1RX218) (d). The adenosine-binding subdomain is colored green for hE–NADPH and pink for hE–NADP+–FOL. Ligands are shown as sticks, with NADPH in green, NADP+ in magenta and FOL in yellow. The adenosine-binding subdomain is colored purple for ecE–NADPH and yellow for ecE–NADP+–FOL, with NADPH in purple, NADP+ in orange and folate in yellow. e,f,g,h Surface representations of hE–NADPH (e), ecE–NADPH (f), hE–NADP+–FOL (g) and ecE–NADP+–FOL (h). Residues highlighting the opening and closing of the active site cleft are colored in red. i,j Difference distance matrix for hE–NADPH and hE–NADP+–FOL (i) and ecE–NADPH and ecE–NADP+–FOL (j), showing the magnitude and character of the conformational changes associated with the hinge motions.