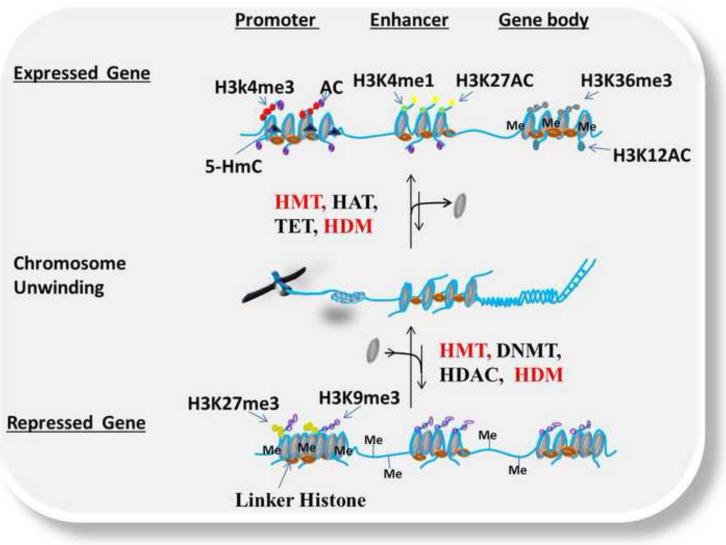
Figure 1.
Dynamic epigenetic regulation in normal cells showing crossed talk between epigenetic modifications resulting in transcriptional activation or repression. This figure depicts different epigenetic modifications under normal conditions for both expressed and repressed genetic targets. Histone Acetyltransferases (HATs), Histone Methyltransferase (HMTs), Ubiquitin ligases (E3), Ten Eleven Translocation (TET) DNA hydroxylase (convert 5mC to 5HmC) and Kinases add transcriptionally active epigenetic marks (e.g. AC, H3K4me3, H3K4me1, H3K27Ac, H3K36me3, H3K12Ac and 5HmC) to promoters, enhancers and gene bodies which promote the formation of euchromatin and transcriptional activation (Top). DNA Methyltransferease (DNMT), Histone Methyltransferase (HMT), Ubiquitin ligase (E3), Histone Deacetyltransferase (HDAC) put repressive epigenetic marks (H3K27me3, H3K9me3 and DNA methylation) to promoters, enhancers and gene bodies promoting the formation of heterochromatin and transcriptional repression (Bottom).