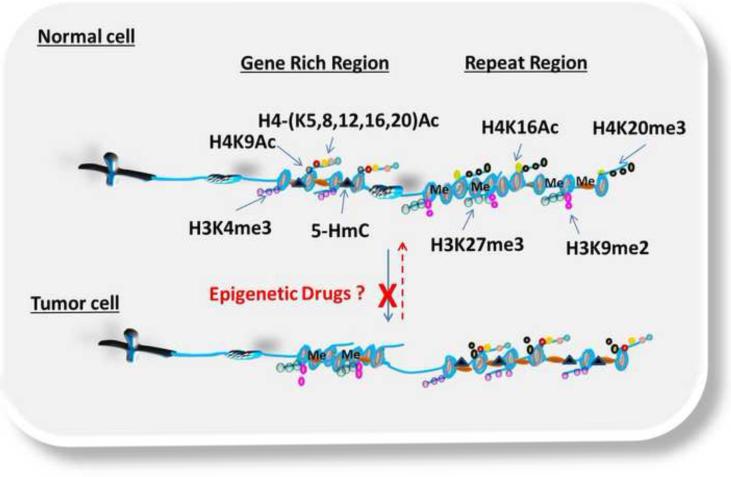
Figure 2.
Simplified version of the dynamic plasticity of epigenetic modifications in normal and tumor cells. In normal cells, gene rich regions are dynamically enriched with active transcription marks (i.e. H4 acetylation at lysine-5,-8,-12 and −16 (H4-(K5,8,12,16)Ac), H4 acetylation at lysine-9 (H4K9Ac), H3 trimethylation at lysine-4 (H3K4me3), 5-hydroxymethylcytosine (5-HmC)) to allow the transcription of tumor suppressor genes, while repeat regions are enriched with repressive marks (H4 acetylation at lysine 16 (H4K16Ac), H4 trimethylation at lysine-20 (H3K20me3), H3 trimethylation at lysine-27(H3K27me3), H3 dimethylation at lysine-9(H3K9me2) and DNA methylation (Me)) that lead to heterochromatin formation. In tumor cells, this dynamic is reversed leading to increased repressive marks at gene rich region and active marks at repeat regions. The dotted red arrow denotes the reversal of these epigenetic modifications by epigenetic drugs to the original state or to an alternate intermediate state. The red X denotes blockage of aberrant epigenetic modification by epigenetic drugs.