Figure 2.
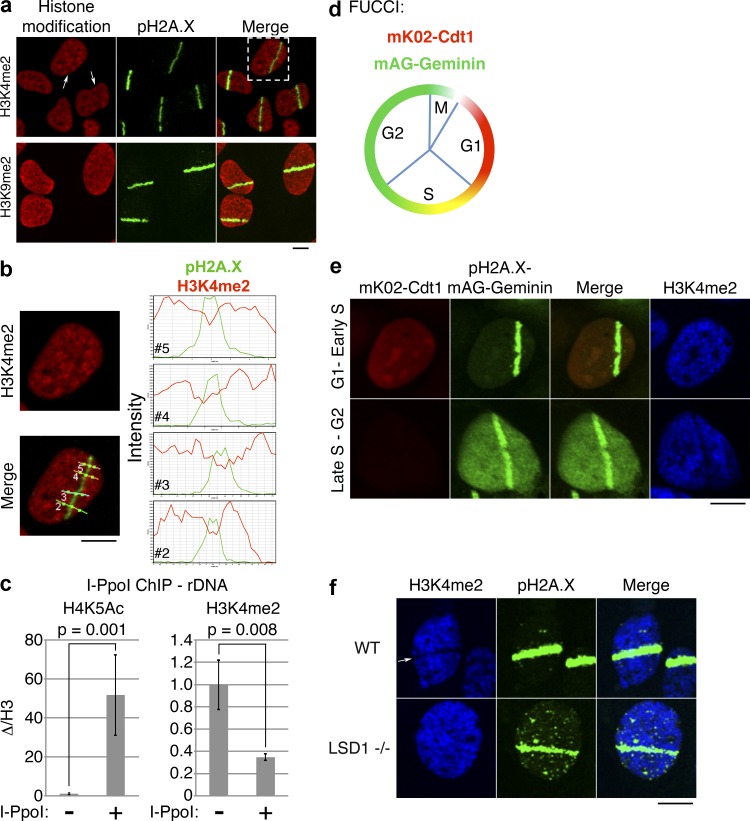
H3K4me2 demethylation marks sites of DNA damage and is cell cycle and LSD1 dependent. (a) UV laser microirradiation was performed on U2OS cells, and the cells were stained with antibodies against the indicated histone modifications. Arrows indicate nuclei with apparent loss of H3K4me2 signal. (b) Magnified view of the cell from top right of panel a, showing H3K4me2 or merged H3K4me2/pH2A.X signals. Intensity profiles of H3K4me2 (red) and pH2A.X (green) signals through the four indicated lines (2–5) are shown on the right. (c) I-PpoI ChIP/qPCR was performed using the indicated histone modification-specific antibodies and primers specific for rDNA. Error bars represent ± standard error. (d) Schematic of FUCCI cells. FUCCI cells coexpress a fragment of Cdt1 linked to the fluorescent protein mK02 (monomeric Kusabira orange 2), as well as a fragment of Geminin linked to the fluorescent protein mAG (monomeric Azami green). (e) UV laser microirradiation was performed on U2OS-FUCCI cells and subsequently stained for pH2A.X and H3K4me2. (f) UV laser microirradiation was performed on wild-type (WT) and LSD1−/− MEFs and subsequently stained for the indicated histone modifications. Bars, 10 µm.