Figure 1.
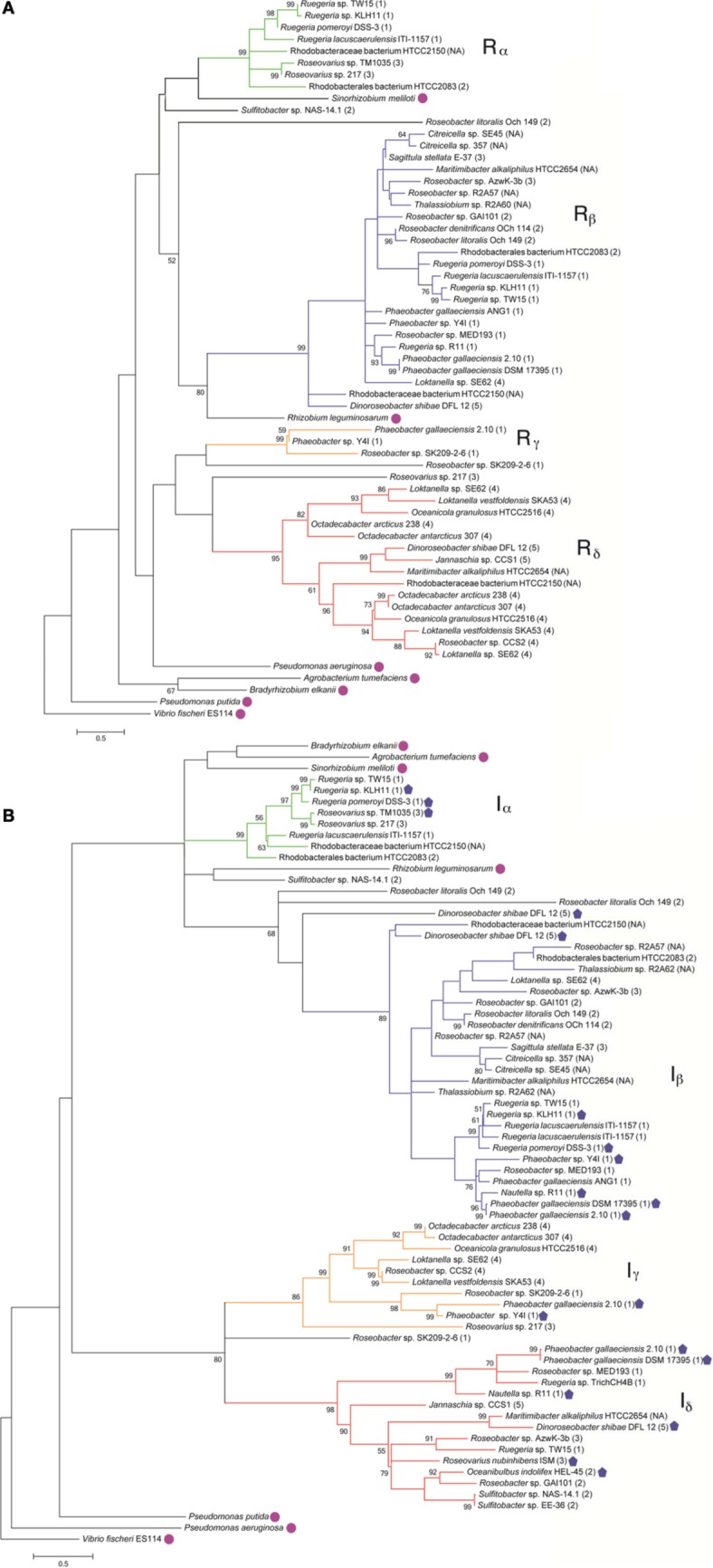
Maximum likelihood phylogenetic trees of Roseobacter LuxR- (A) and LuxI-like (B) deduced amino acid sequences (see Appendix for details). Strain designations are shown and gene locus tags of the corresponding gene sequences can be found in Table A1. The scale bar represents the substitutions per sequence position. The Roseobacter clade number is represented in parentheses after the organism name and follows the classification system identified in Newton et al., 2010. Proposed designations of LuxR and LuxI subgroups in roseobacters are indicated by Greek character subscript and color. Bootstrap values <50% (from 1000 iterations) are shown at branch nodes. Sequences designated with a closed pentagon indicate organisms that have been shown experimentally, by either bioreporters or mass spectrometry, to produce AHLs (Wagner-Dobler et al., 2005; Rao et al., 2006; Bruhn et al., 2007; Berger et al., 2011; Case et al., 2011; Zan et al., 2012). Sequences designated with a circle are non-roseobacters.
