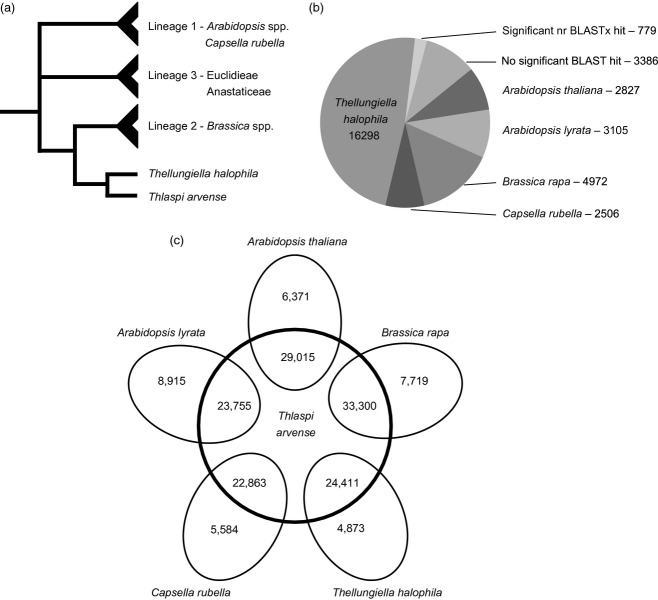
Figure 2.
Comparative transcriptomics of pennycress versus five Brassicaceae species.
(a) Representation of the Brassicaceae phylogeny, adapted from Beilstein et al. (2010) and Franzke et al. (2011).
(b) BLASTx comparison of the pennycress transcriptome assembly versus A. thaliana, Arabidopsis lyrata, Brassica rapa, Capsella rubella and Thellungiella halophila. The top blast hit (e ≤ 0.05) for each pennycress transcript versus the five species is shown. Contigs without significant hits were then compared to the NCBI peptide non-redundant database.
(c) Five pairwise tBLASTn comparisons of Brassicaceae species to the pennycress transcriptome assembly. Sequences with significant homology (e ≤ 0.05 and positive match percentage ≧70%) shared between the five Brassicaceae species and pennycress (Thlaspi arvense) are shown in the inner circle.