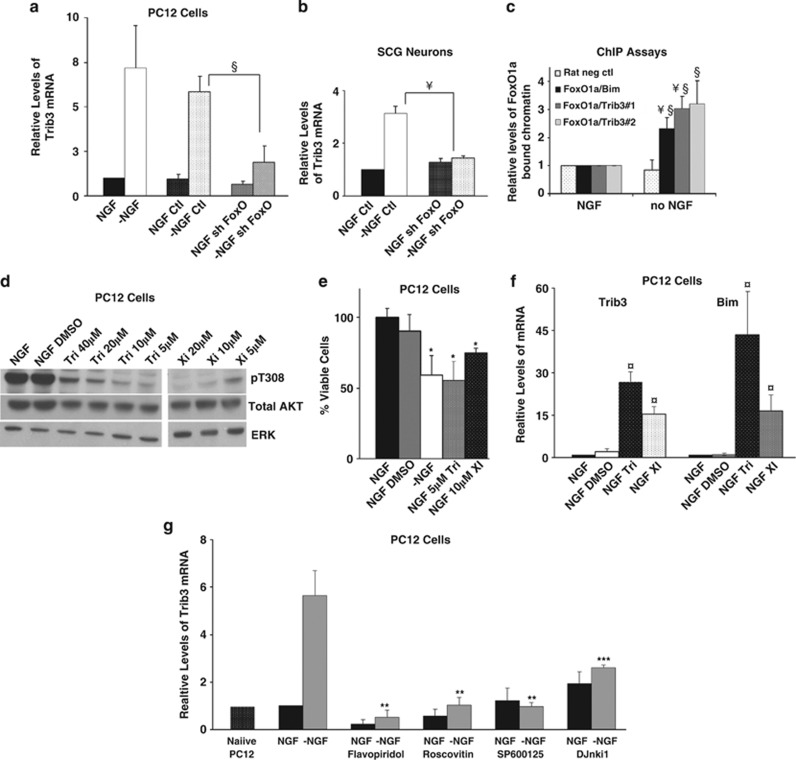
Figure 8.
Regulation of Trib3 expression. (a and b) Induction of Trib3 in response to NGF deprivation requires FoxO. Neuronal PC12 cells (a) and SCG neurons (b) were infected with shFoxO or control shRNA expressing lentivirus for 72 h. Cultures were maintained with or without NGF for an additional 16 h for PC12 cells and 24 h for SCG neurons. Total mRNA was isolated and quantitative PCR was performed using Trib3 primers. Rat α-tubulin mRNA levels were used to normalize input cDNA. Values are means±S.E. of data from three independent experiments (a) and one experiment (b) each done in triplicate; §P<0.05; ¥P<0.005, (Student's t-test) compared with −NGF control. (c) ChIP assay indicates that FoxO1a occupancy of rat Trib3 promoter regions increases after NGF deprivation. Neuronal PC12 cells were either maintained with NGF or subjected to NGF withdrawal for 16 h. After crosslinking, the chromatin was sheared and immunoprecipitated using antibody against total FoxO1a or rabbit immunoglobulin G (IgG) isotype control. After reversing the crosslink, the genomic DNA was purified and subjected to quantitative PCR using two different primer sets designed to amplify promoter regions of Trib3 identified as corresponding to the putative FoxO1a binding sites. FoxO1a/Trib3#1 and FoxO1a/Trib3#2 refer to primer sets designed to amplify a region near the FoxO1a DNA located approximately 700 and 2000 bp upstream of the rat Trib3 translation initiation site, respectively, and that include the putative FoxO1a binding sites. Primers, including a previously identified FoxO binding site in the Bim promoter, were used as a positive control (FoxO1a/Bim) and commercially made primers recognizing a region of a ‘gene desert' on rat chromosome 3 were used as a negative control (‘rat neg control') for quantitative PCR. QPCR signals were normalized as bound/INPUT, and background signals obtained from rabbit IgG immunoprecipitated samples (25–50% of the values obtained with NGF and FoxO1a antibody) were subtracted. The data are normalized against NGF control samples for all primer sets and are presented as mean relative levels of FoxO1a-enriched chromatin in cells±S.E. of three independent experiments each conducted in triplicate. §P<0.05; ¥P<0.005, (Student's t-test) compared with NGF control and §P<0.05 compared with no NGF/rat negative control primers. (d) Inhibition of Akt phosphorylation by various inhibitors. Cell lysates of neuronal PC12 cells treated with various concentrations of Triciribine, and compound XI in the presence of NGF for 20 h were subjected to SDS-PAGE and immunoblotted using antisera against phospho-Akt (pT308), total Akt and ERK. (e) Inhibition of Akt activity promotes cell death. Neuronal PC12 cells were treated with the indicated concentrations of Akt inhibiting drugs for 20 h while maintained with NGF. Viable cells were then counted. Values are means±S.E. of three independent experiments each performed in triplicate. *P<0.05 (Student's t-test), compared with NGF/DMSO (dimethyl sulfoxide). (f) Akt inhibition induces Trib3 mRNA. cDNA derived from cultures treated as in panel (e) were subjected to quantitative PCR using Trib3 and Bim primers. Rat α-tubulin mRNA levels were used to normalize input cDNA. Values are means±S.E. of three independent experiments each performed in triplicate. ¤P<0.005, (Student's t-test) compared with NGF/DMSO. (g) Inhibitors of JNK and cyclin-dependent kinases suppress Trib3 mRNA induction in response to NGF deprivation. Neuronal PC12 cells were treated with the Cdk inhibiting drugs roscovitine and flavopiridol and the JNK inhibitors SP600125 and DJnkI1 at 10 μM each (g) in the presence and absence of NGF for 16 h. cDNA derived from the cultures was subjected to quantitative PCR using Trib3 primers. Rat α-tubulin mRNA levels were used to normalize input cDNA. Values are means±S.E. of four independent experiments performed in triplicate, **P<0.05 and ***P<0.05 of two independent experiments performed in triplicate (Student's t-test) compared with −NGF control